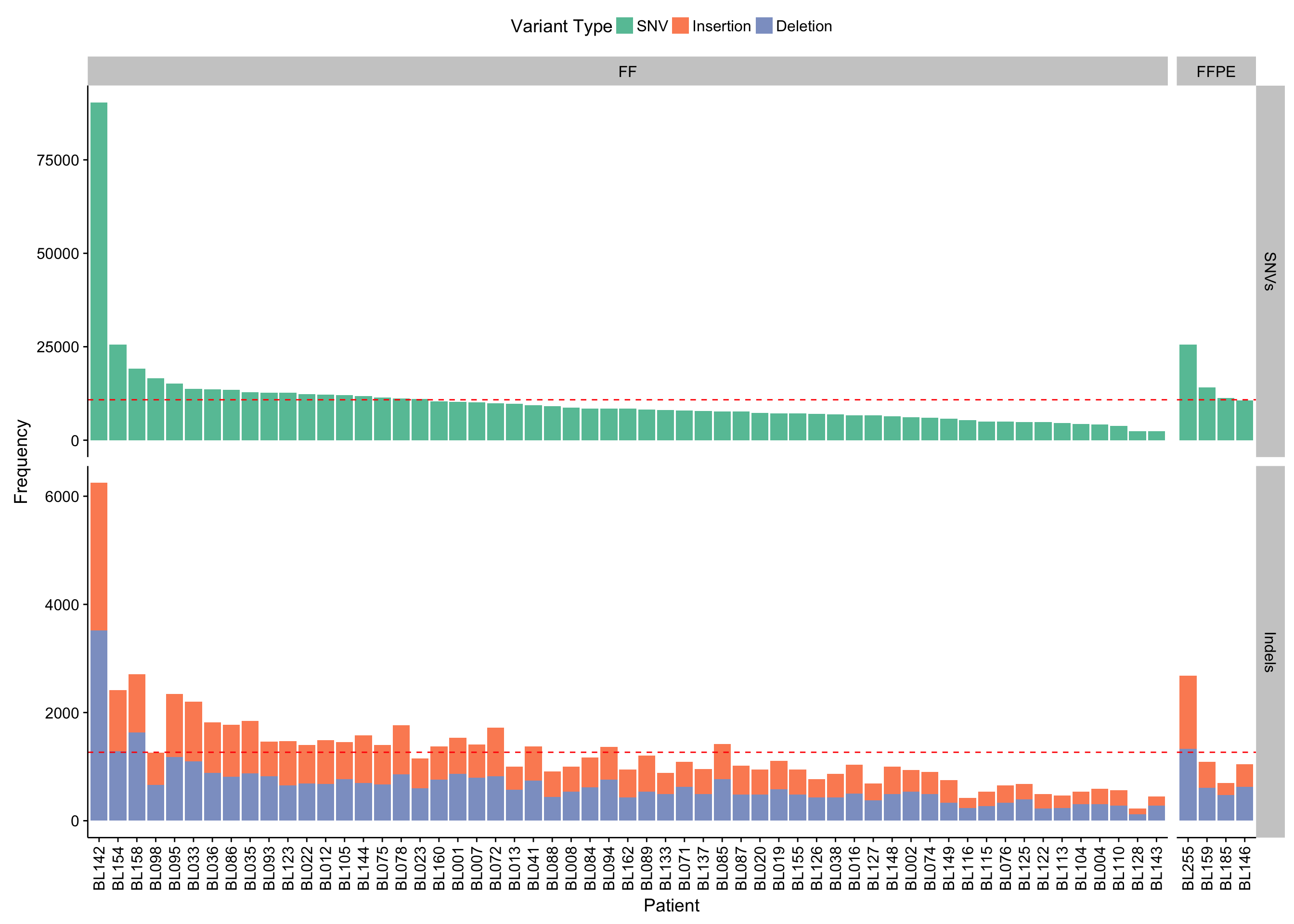
barplot_maf_mutfreq_data <-
maf@data %>%
dplyr::count(patient, ff_or_ffpe, Variant_Type) %>%
dplyr::ungroup() %>%
tidyr::spread(Variant_Type, n) %>%
dplyr::mutate(patient = forcats::fct_reorder(patient, -SNP)) %>%
tidyr::gather(Variant_Type, n, -patient, -ff_or_ffpe) %>%
dplyr::mutate(
Variant_Type = dplyr::recode_factor(
Variant_Type, SNP = "SNV", INS = "Insertion", DEL = "Deletion"),
Variant_Class = dplyr::recode_factor(
Variant_Type, SNV = "SNVs", Insertion = "Indels", Deletion = "Indels"))
barplot_maf_mutfreq_stats <-
barplot_maf_mutfreq_data %>%
dplyr::group_by(Variant_Class, patient) %>%
dplyr::summarise(n = sum(n)) %>%
dplyr::summarise(n_patients = dplyr::n_distinct(patient),
max = max(n),
mean = round(mean(n)))
barplot_mut_load <-
ggplot(barplot_maf_mutfreq_data) +
geom_col(aes(x = patient, y = n, fill = Variant_Type)) +
geom_hline(data = barplot_maf_mutfreq_stats, aes(yintercept = mean),
colour = "red", linetype = 2) +
facet_grid(Variant_Class ~ ff_or_ffpe, scales = "free", space = "free_x") +
scale_fill_brewer(palette = "Set2") +
theme(legend.position = "top") +
rotate_x_text() +
labs(x = "Patient", y = "Frequency", fill = "Variant Type")
barplot_mut_load
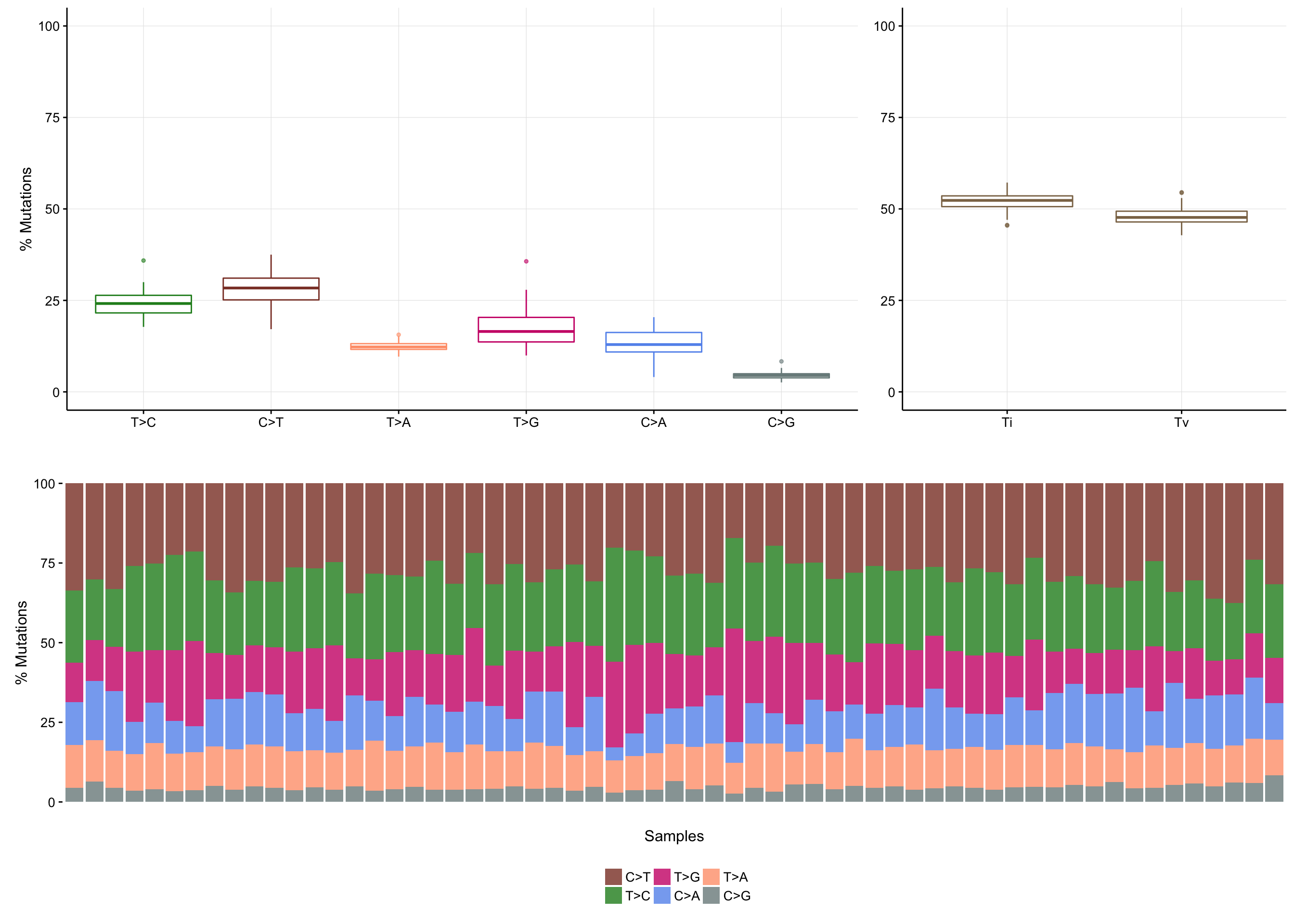
maf_titv <- titv(maf, useSyn = TRUE, plot = FALSE)
plotTiTv(maf_titv)
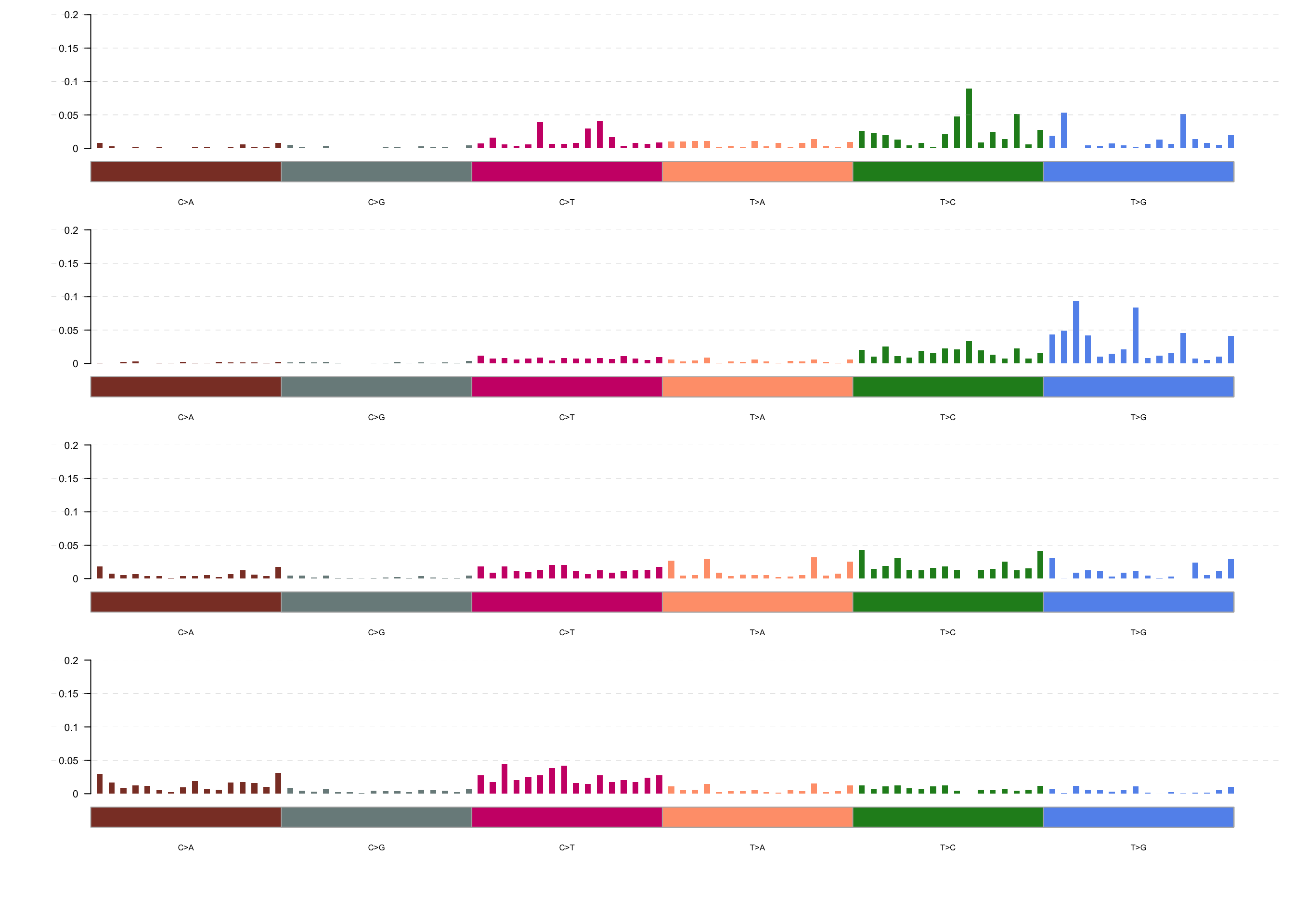
maf_tnm <- trinucleotideMatrix(maf, paths$genome, useSyn = TRUE)
maf_sig <- extractSignatures(maf_tnm, nTry = 6, plotBestFitRes = FALSE)
method seed rng metric rank sparseness.basis sparseness.coef rss evar
1: brunet random 2 KL 2 0.3095876 0.4142273 10956186 0.9654932
2: brunet random 3 KL 3 0.3552028 0.3575419 3364764 0.9894026
3: brunet random 4 KL 4 0.3646364 0.3033678 2216414 0.9930194
4: brunet random 1 KL 5 0.3728185 0.2633642 1827725 0.9942435
5: brunet random 3 KL 6 0.3598597 0.2990961 1553944 0.9951058
silhouette.coef silhouette.basis residuals niter cpu cpu.all nrun cophenetic dispersion
1: 1.0000000 1.0000000 15529.722 440 0.121 13.547 10 0.9740442 0.8784413
2: 0.8378465 0.8018052 10604.867 740 0.154 13.575 10 0.9828152 0.8398495
3: 0.6227660 0.6963675 8463.816 1040 0.225 13.953 10 0.9826599 0.7913249
4: 0.4967904 0.5980047 7337.924 920 0.222 14.313 10 0.9500396 0.6964472
5: 0.5434427 0.4506834 6552.669 1100 0.298 14.911 10 0.9657381 0.7752647
silhouette.consensus
1: 0.9561801
2: 0.8924888
3: 0.8574604
4: 0.6488602
5: 0.7396883
corrplot::corrplot(maf_sig$coSineSimMat, is.corr = FALSE, tl.cex = 0.6,
tl.col = 'black', cl.cex = 0.6)

maf_sig$contributions %>%
set_colnames(get_patient_id(colnames(.))) %>%
plot_heatmap(colours, gannotations, cutree_cols = 3, cluster_rows = FALSE)

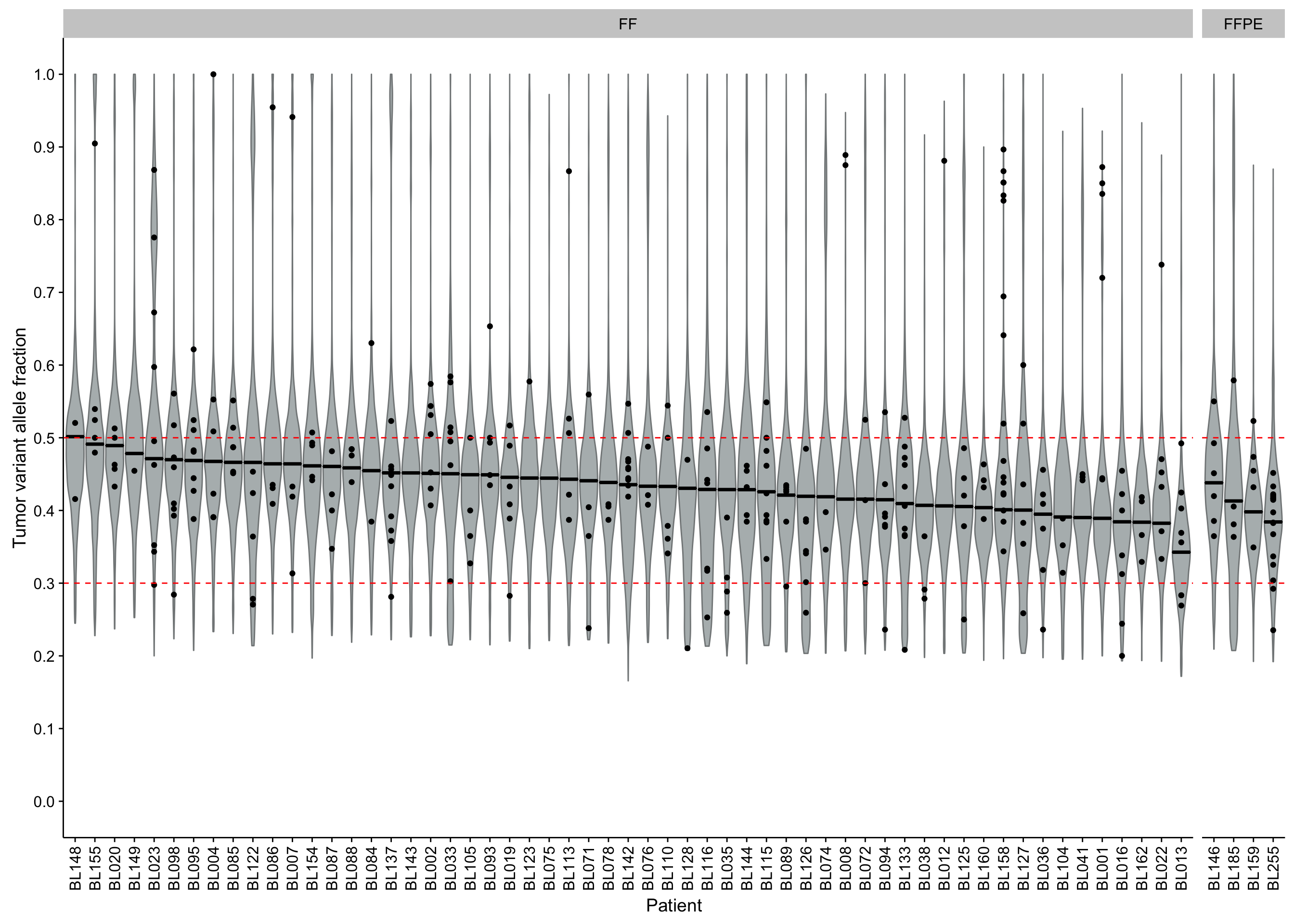
violinplot_maf_vaf <-
maf@data %>%
mutate(
patient = fct_reorder(patient, -vaf_peak),
is_bl_gene = Hugo_Symbol %in% genes$bl & is_nonsynonymous(Consequence)) %>% {
ggplot(., aes(patient, t_vaf)) +
geom_violin(scale = "width", fill = "#B4BABB", color = "#838788") +
geom_point(data = filter(., is_bl_gene)) +
geom_crossbar(
data = group_by(., patient) %>% slice(1),
aes(x = patient, y = vaf_peak, ymin = vaf_peak, ymax = vaf_peak)) +
geom_hline(yintercept = c(0.3, 0.5), color = "red", linetype = 2) +
scale_y_continuous(breaks = seq(0, 1, 0.1), limits = c(0, 1)) +
facet_grid(~ff_or_ffpe, scales = "free_x", space = "free_x") +
rotate_x_text() +
labs(x = "Patient", y = "Tumor variant allele fraction")}
violinplot_maf_vaf
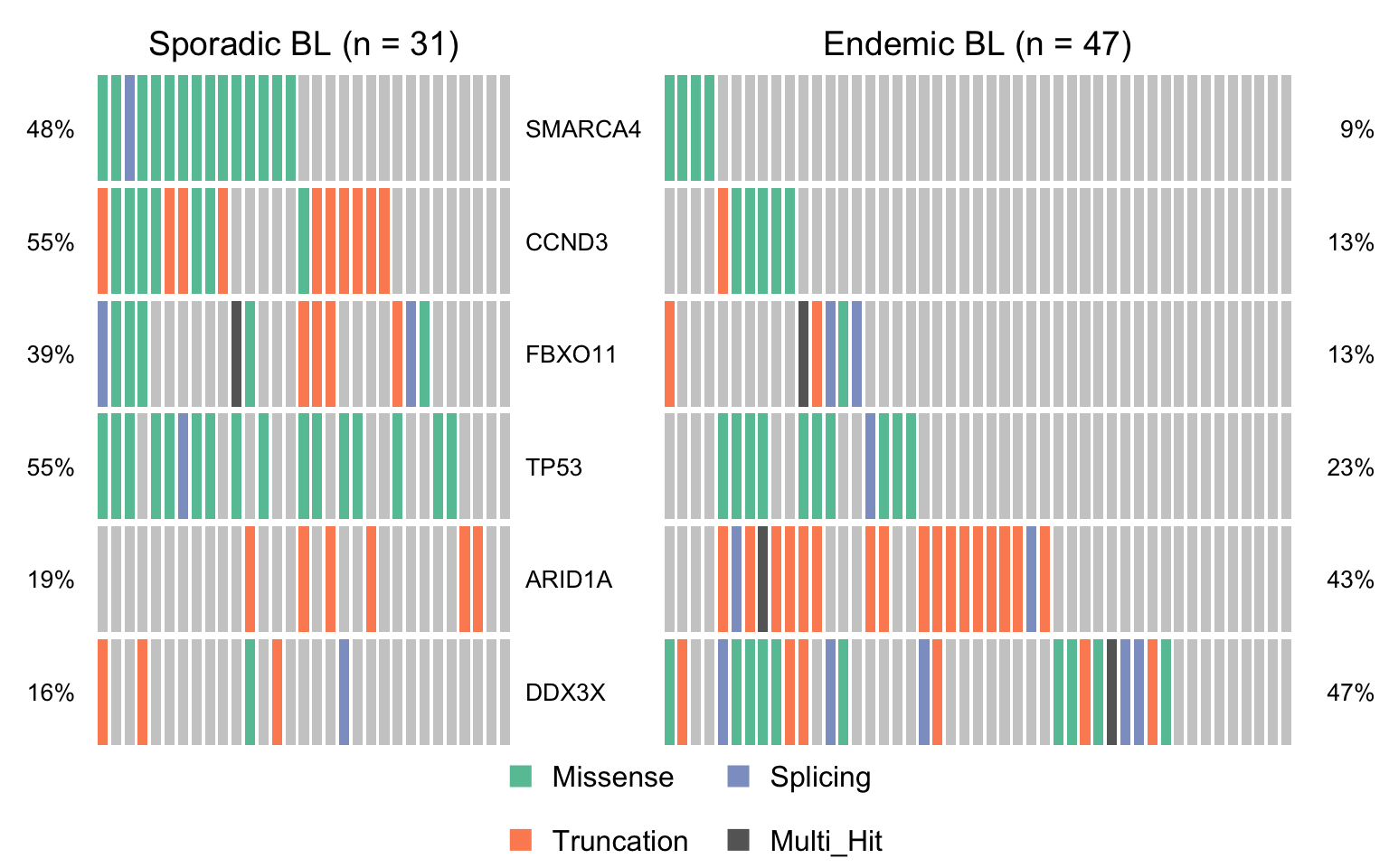
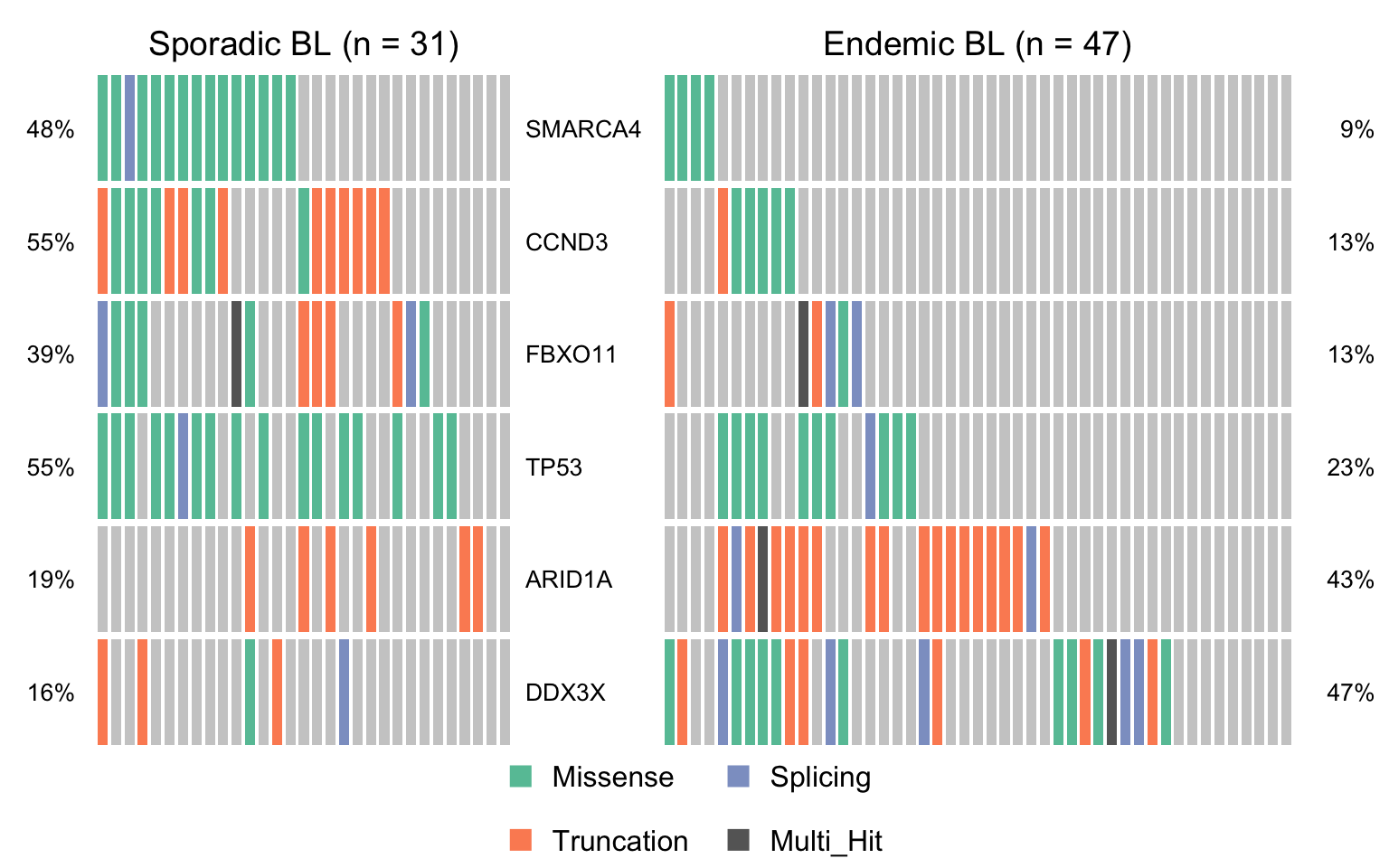
maf_ebl <- subsetMaf(mmaf, query = "clinical_variant == 'Endemic'",
includeSyn = TRUE, mafObj = TRUE)
maf_sbl <- subsetMaf(mmaf, query = "clinical_variant == 'Sporadic'",
includeSyn = TRUE, mafObj = TRUE)
maf_ebl_vs_sbl <- mafCompare(maf_ebl, maf_sbl, 'Endemic BL', 'Sporadic BL', minMut = 4)
sig_genes_ebv_vs_sbl <-
maf_ebl_vs_sbl$results %>%
filter(Hugo_Symbol %in% smgs) %$%
Hugo_Symbol[pval < 0.05][order(or[pval < 0.05])]
if (length(sig_genes_ebv_vs_sbl) > 0) {
cooncoplot_maf_ebl_vs_sbl <- coOncoplot(
maf_sbl, maf_ebl, sig_genes_ebv_vs_sbl,
m1Name = 'Sporadic BL', m2Name = 'Endemic BL',
removeNonMutated = FALSE, colors = colours$categs)
print(cooncoplot_maf_ebl_vs_sbl)
}
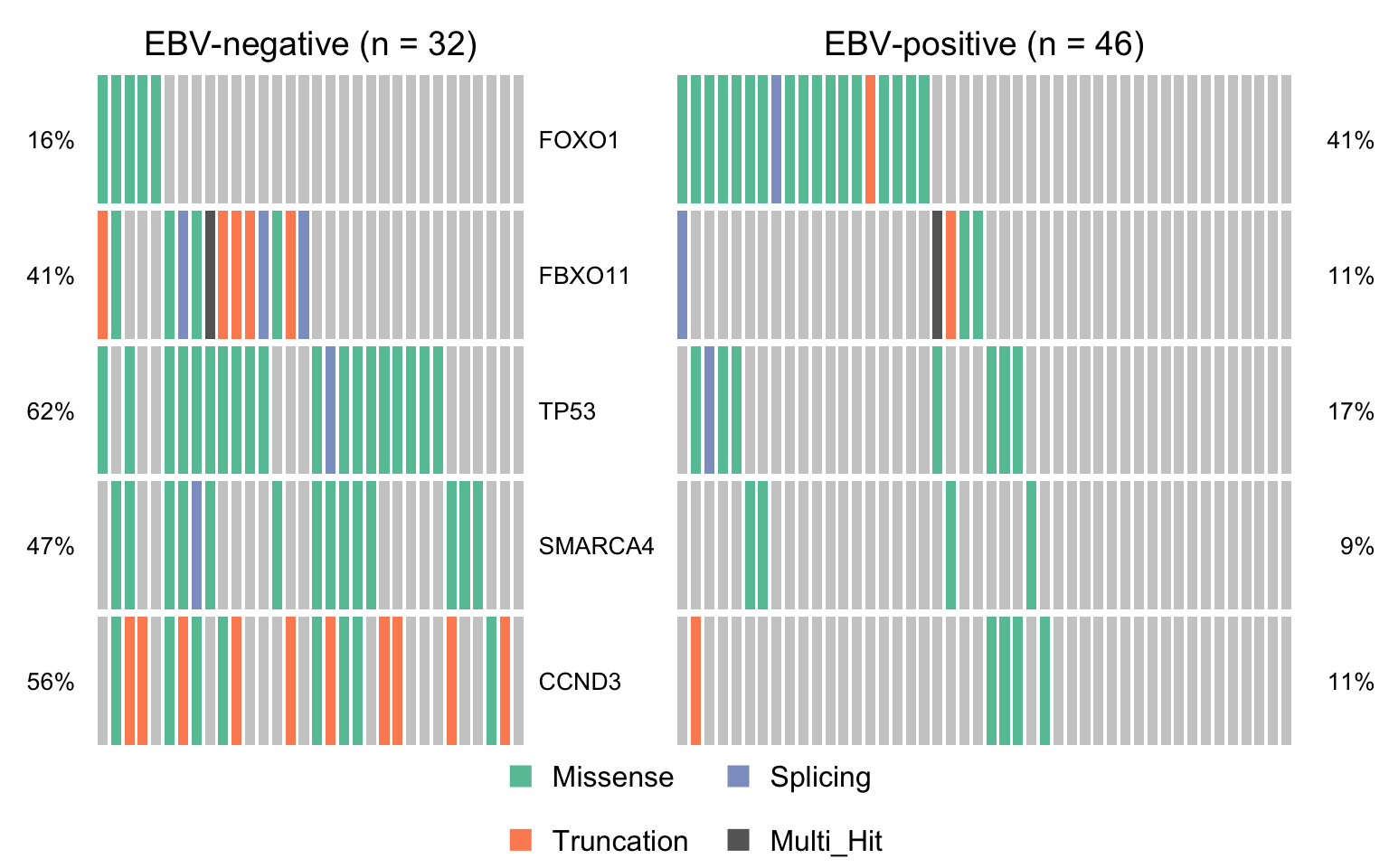
maf_ebvneg <- subsetMaf(mmaf, query = "ebv_type == 'None'",
includeSyn = TRUE, mafObj = TRUE)
maf_ebvpos <- subsetMaf(mmaf, query = "ebv_type != 'None'",
includeSyn = TRUE, mafObj = TRUE)
maf_ebvneg_vs_ebvpos <- mafCompare(maf_ebvneg, maf_ebvpos, 'EBV-negative', 'EBV-positive', minMut = 4)
sig_genes_ebvneg_vs_ebvpos <-
maf_ebvneg_vs_ebvpos$results %>%
filter(Hugo_Symbol %in% smgs) %$%
Hugo_Symbol[pval < 0.05][order(or[pval < 0.05])]
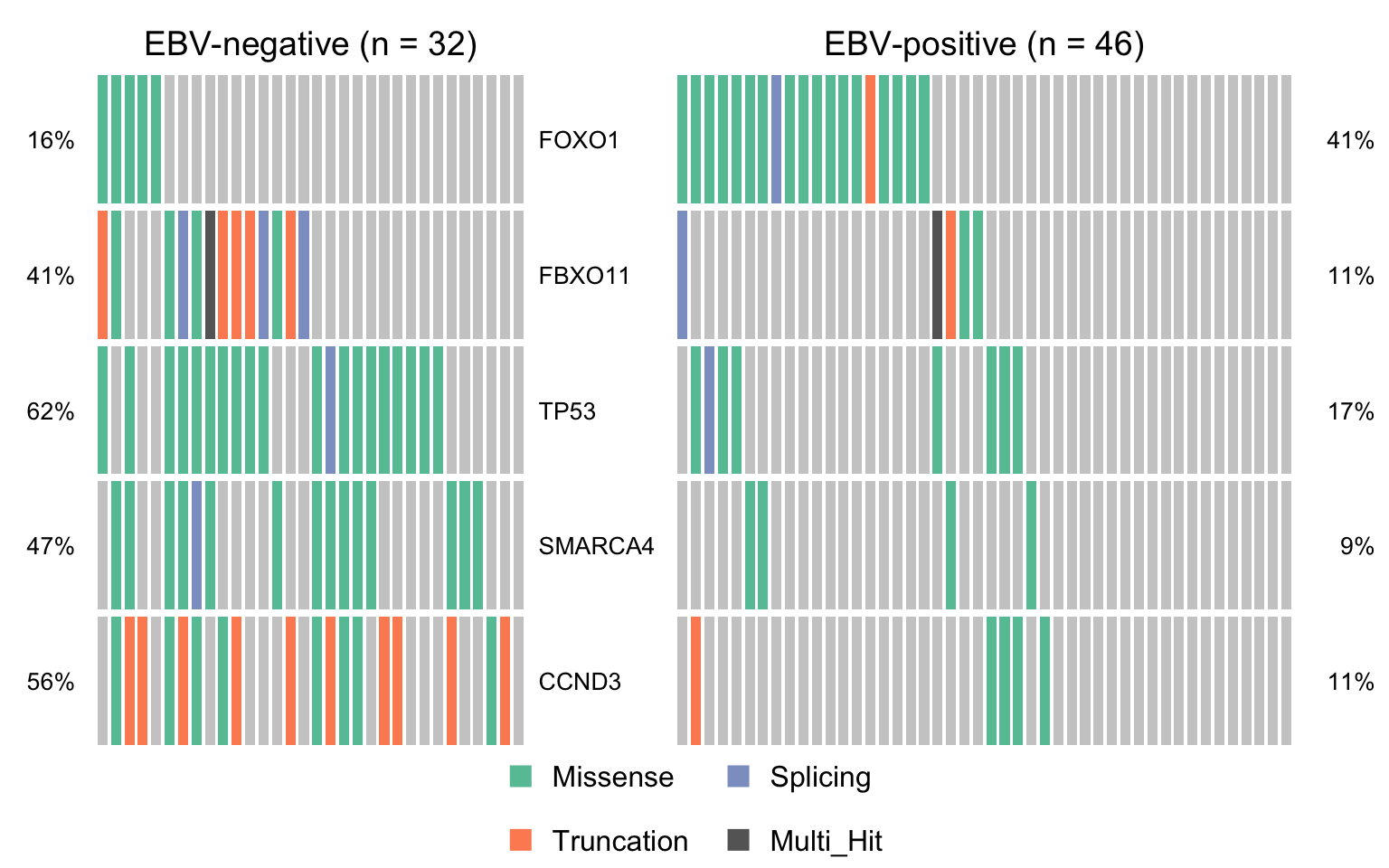
if (length(sig_genes_ebvneg_vs_ebvpos) > 0) {
cooncoplot_maf_ebvneg_vs_ebvpos <- coOncoplot(
maf_ebvneg, maf_ebvpos, sig_genes_ebvneg_vs_ebvpos,
m1Name = 'EBV-negative', m2Name = 'EBV-positive',
removeNonMutated = FALSE, colors = colours$categs)
print(cooncoplot_maf_ebvneg_vs_ebvpos)
}
maf_ebv1 <- subsetMaf(mmaf, query = "ebv_type == 'Type 1'",
includeSyn = TRUE, mafObj = TRUE)
maf_ebv2 <- subsetMaf(mmaf, query = "ebv_type == 'Type 2'",
includeSyn = TRUE, mafObj = TRUE)
maf_ebv1_vs_ebv2 <- mafCompare(maf_ebv1, maf_ebv2, 'EBV Type 1', 'EBV Type 2', minMut = 4)
sig_genes_ebv1_vs_ebv2 <-
maf_ebv1_vs_ebv2$results %>%
filter(Hugo_Symbol %in% smgs) %$%
Hugo_Symbol[pval < 0.05][order(or[pval < 0.05])]
if (length(sig_genes_ebv1_vs_ebv2) > 0) {
cooncoplot_maf_ebv1_vs_ebv2 <- coOncoplot(
maf_ebv1, maf_ebv2, c(sig_genes_ebv1_vs_ebv2, "TP53"),
m1Name = 'EBV Type 1', m2Name = 'EBV Type 2',
removeNonMutated = FALSE, colors = colours$categs)
print(cooncoplot_maf_ebv1_vs_ebv2)
}
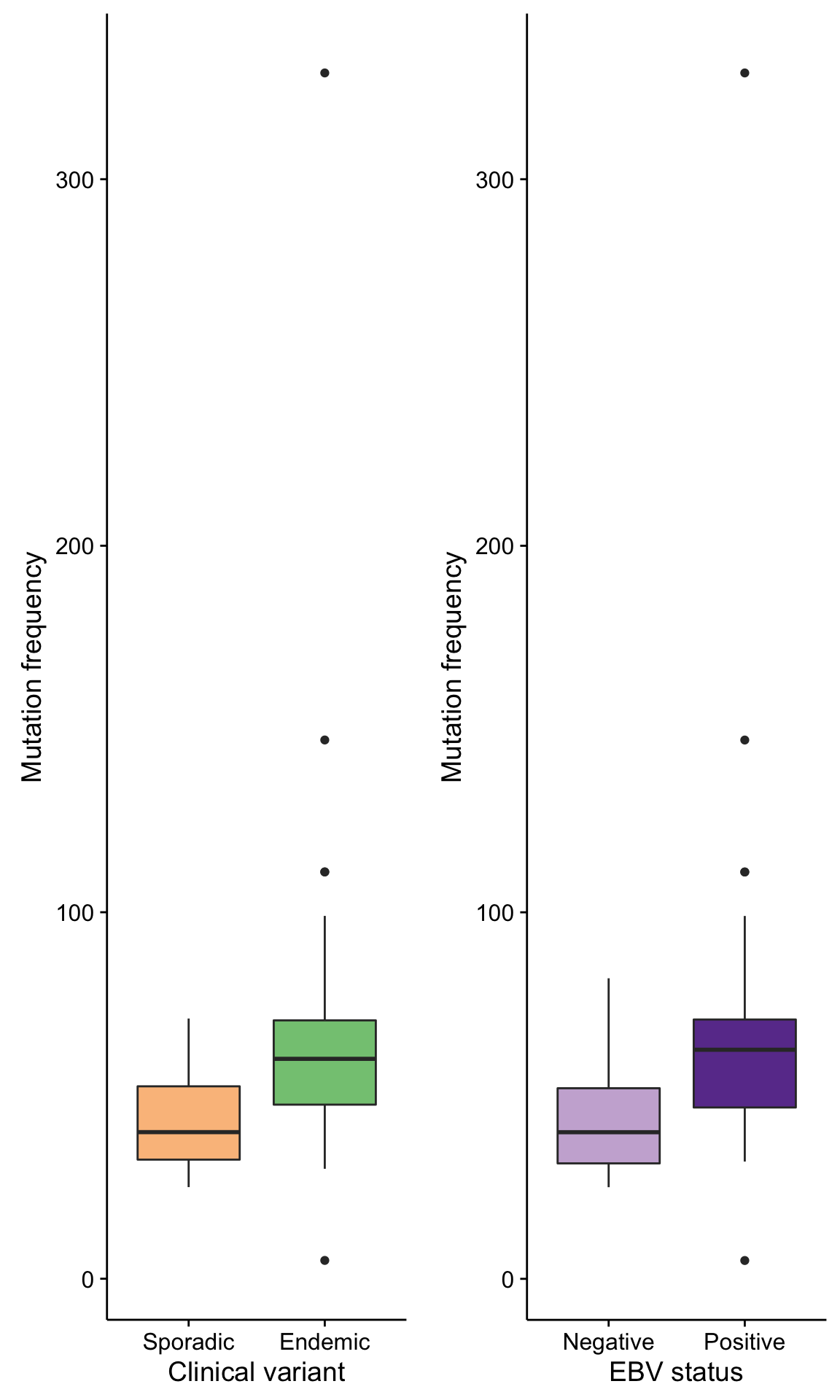
boxplot_maf_compare_all_mutfreq_cv <-
mmaf@data %>%
count(clinical_variant, Tumor_Sample_Barcode) %>%
ggplot(aes(clinical_variant, n, fill = clinical_variant)) +
geom_boxplot() +
scale_fill_manual(values = colours$clinical_variant, breaks = NULL) +
labs(x = "Clinical variant", y = "Mutation frequency")
boxplot_maf_compare_all_mutfreq_ebv <-
mmaf@data %>%
count(ebv_status = ifelse(ebv_type == "None", "Negative", "Positive"), Tumor_Sample_Barcode) %>%
ggplot(aes(ebv_status, n, fill = ebv_status)) +
geom_boxplot() +
scale_fill_manual(values = colours$ebv_status, breaks = NULL) +
labs(x = "EBV status", y = "Mutation frequency")
gridExtra::grid.arrange(
boxplot_maf_compare_all_mutfreq_cv, boxplot_maf_compare_all_mutfreq_ebv, ncol = 2)
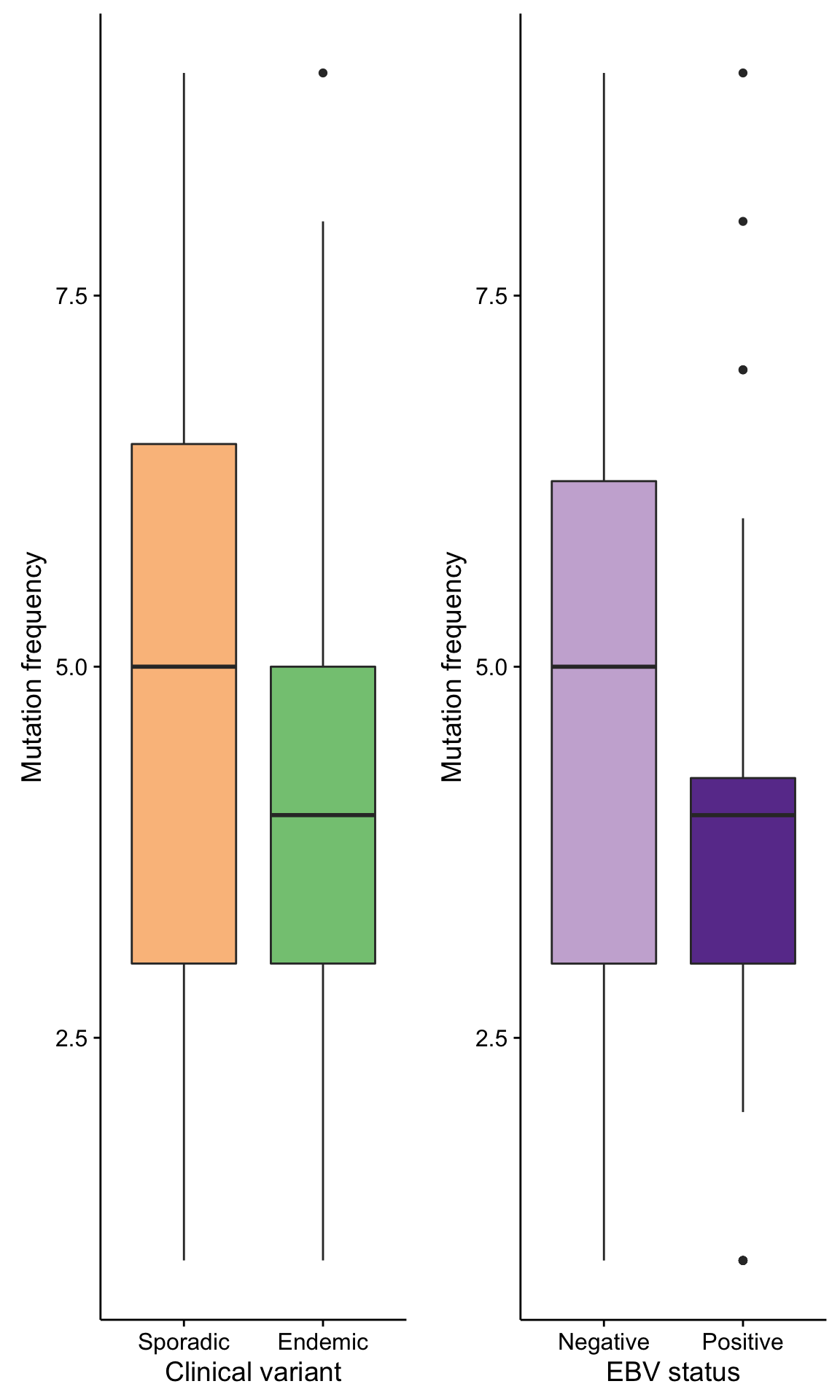
boxplot_maf_compare_nonsyn_smg_mutfreq_cv <-
mmaf@data %>%
filter(is_nonsynonymous(Consequence), Hugo_Symbol %in% smgs) %>%
count(clinical_variant, Tumor_Sample_Barcode) %>%
ggplot(aes(clinical_variant, n, fill = clinical_variant)) +
geom_boxplot() +
scale_fill_manual(values = colours$clinical_variant, breaks = NULL) +
labs(x = "Clinical variant", y = "Mutation frequency")
boxplot_maf_compare_nonsyn_smg_mutfreq_ebv <-
mmaf@data %>%
filter(is_nonsynonymous(Consequence), Hugo_Symbol %in% smgs) %>%
count(ebv_status = ifelse(ebv_type == "None", "Negative", "Positive"), Tumor_Sample_Barcode) %>%
ggplot(aes(ebv_status, n, fill = ebv_status)) +
geom_boxplot() +
scale_fill_manual(values = colours$ebv_status, breaks = NULL) +
labs(x = "EBV status", y = "Mutation frequency")
gridExtra::grid.arrange(
boxplot_maf_compare_nonsyn_smg_mutfreq_cv, boxplot_maf_compare_nonsyn_smg_mutfreq_ebv, ncol = 2)
ig_loci <- GRanges(
c("chr2", "chr8", "chr14", "chr22"),
IRanges(c(88700000, 127735000, 105500000, 22600000),
c(89300000, 127741000, 107000000, 23000000)))
chroms <- c(paste0("chr", 1:22), "chrX")
seqlens <- seqlengths(BSgenome.Hsapiens.UCSC.hg38::Hsapiens)[chroms]
gen <- data.frame(chroms, seqlens) %>% set_colnames(c("V1", "V2"))
# gaps_gr <-
# read_tsv_quiet(paths$gaps) %>%
# rename(seqnames = `#chrom`, start = chromStart, end = chromEnd) %>%
# makeGRangesFromDataFrame()
bins <-
tileGenome(seqlens, tilewidth = 5000, cut.last.tile.in.chrom = TRUE) %>%
subsetByOverlaps(ig_loci, invert = TRUE)
# maf_grl <-
# maf@data %>%
# as.data.frame() %>%
# # filter_nonsyn(inverse = TRUE) %>%
# makeGRangesListFromDataFrame(
# split.field = "biospecimen_id",
# seqnames.field = "Chromosome",
# start.field = "Start_Position",
# end.field = "End_Position")
#
# mut_counts <-
# map(as.list(maf_grl), ~countOverlaps(bins, .x)) %>%
# # map(pmin, 1) %>%
# invoke(cbind, .) %>%
# rowSums()
#
# map(as.list(maf_grl), ~countOverlaps(bins, .x)) %>%
# invoke(cbind, .) %>%
# as.data.frame() %>%
# write_tsv("~/Desktop/counts.10kb_bins.all_muts.no_gaps.tsv")
#
# mut_counts_fit <- MASS::fitdistr(mut_counts, "Poisson")
#
# mut_counts_df <-
# bins %>%
# as.data.frame() %>%
# inset("count", value = mut_counts) %>%
# select(seqnames, start, end, count) %>%
# mutate(
# pval = ppois(count, mut_counts_fit$estimate, lower.tail = FALSE),
# qval = p.adjust(pval, "BH"),
# nlog_pval = pmin(-log10(pval), 20),
# signif = qval < 0.00001)
#
# mut_counts_df %>%
# filter(count > 2) %>%
# ggplot(aes(start, nlog_pval, color = seqnames)) +
# geom_point() +
# scale_x_continuous(breaks = NULL) +
# scale_colour_discrete(breaks = NULL) +
# facet_grid(~seqnames, scales = "free_x", space = "free_x") +
# theme(
# panel.background = element_rect(fill = "grey95"),
# panel.spacing = unit(0.2, "lines"))
mut_counts_path <- project("etc/noncoding_pvals.tsv")
mut_counts_cols <- c("row_id", "binSum", "PoissonPvalue", "lognormalPvalue", "binomialPvalue")
mut_counts_df <-
bins %>%
as.data.frame() %>%
bind_cols(read_tsv_quiet(mut_counts_path, col_names = mut_counts_cols, skip = 1)) %>%
mutate(nlog_pval = -log(binomialPvalue))
mut_counts_df
seqnames start end width strand row_id binSum PoissonPvalue lognormalPvalue
1 chr1 1 5000 5000 * 1 0 4.256839e-01 7.718511e-01
2 chr1 5001 10000 5000 * 2 0 4.256839e-01 7.718511e-01
3 chr1 10001 15000 5000 * 3 1 1.071823e-01 2.066926e-01
4 chr1 15001 20000 5000 * 4 0 4.256839e-01 7.718511e-01
5 chr1 20001 25000 5000 * 5 0 4.256839e-01 7.718511e-01
6 chr1 25001 30000 5000 * 6 0 4.256839e-01 7.718511e-01
7 chr1 30001 35000 5000 * 7 0 4.256839e-01 7.718511e-01
8 chr1 35001 40000 5000 * 8 0 4.256839e-01 7.718511e-01
9 chr1 40001 45000 5000 * 9 0 4.256839e-01 7.718511e-01
10 chr1 45001 50000 5000 * 10 0 4.256839e-01 7.718511e-01
11 chr1 50001 55000 5000 * 11 0 4.256839e-01 7.718511e-01
12 chr1 55001 60000 5000 * 12 0 4.256839e-01 7.718511e-01
13 chr1 60001 65000 5000 * 13 2 1.886580e-02 4.161957e-02
14 chr1 65001 70000 5000 * 14 0 4.256839e-01 7.718511e-01
15 chr1 70001 75000 5000 * 15 0 4.256839e-01 7.718511e-01
16 chr1 75001 80000 5000 * 16 1 1.071823e-01 2.066926e-01
17 chr1 80001 85000 5000 * 17 0 4.256839e-01 7.718511e-01
18 chr1 85001 90000 5000 * 18 0 4.256839e-01 7.718511e-01
19 chr1 90001 95000 5000 * 19 0 4.256839e-01 7.718511e-01
20 chr1 95001 100000 5000 * 20 0 4.256839e-01 7.718511e-01
21 chr1 100001 105000 5000 * 21 0 4.256839e-01 7.718511e-01
22 chr1 105001 110000 5000 * 22 0 4.256839e-01 7.718511e-01
23 chr1 110001 115000 5000 * 23 0 4.256839e-01 7.718511e-01
24 chr1 115001 120000 5000 * 24 0 4.256839e-01 7.718511e-01
25 chr1 120001 125000 5000 * 25 0 4.256839e-01 7.718511e-01
26 chr1 125001 130000 5000 * 26 0 4.256839e-01 7.718511e-01
27 chr1 130001 135000 5000 * 27 0 4.256839e-01 7.718511e-01
28 chr1 135001 140000 5000 * 28 0 4.256839e-01 7.718511e-01
29 chr1 140001 145000 5000 * 29 0 4.256839e-01 7.718511e-01
30 chr1 145001 150000 5000 * 30 0 4.256839e-01 7.718511e-01
31 chr1 150001 155000 5000 * 31 0 4.256839e-01 7.718511e-01
32 chr1 155001 160000 5000 * 32 0 4.256839e-01 7.718511e-01
33 chr1 160001 165000 5000 * 33 0 4.256839e-01 7.718511e-01
34 chr1 165001 170000 5000 * 34 0 4.256839e-01 7.718511e-01
35 chr1 170001 175000 5000 * 35 0 4.256839e-01 7.718511e-01
36 chr1 175001 180000 5000 * 36 0 4.256839e-01 7.718511e-01
37 chr1 180001 185000 5000 * 37 0 4.256839e-01 7.718511e-01
38 chr1 185001 190000 5000 * 38 0 4.256839e-01 7.718511e-01
39 chr1 190001 195000 5000 * 39 0 4.256839e-01 7.718511e-01
40 chr1 195001 200000 5000 * 40 0 4.256839e-01 7.718511e-01
41 chr1 200001 205000 5000 * 41 0 4.256839e-01 7.718511e-01
42 chr1 205001 210000 5000 * 42 0 4.256839e-01 7.718511e-01
43 chr1 210001 215000 5000 * 43 0 4.256839e-01 7.718511e-01
44 chr1 215001 220000 5000 * 44 0 4.256839e-01 7.718511e-01
45 chr1 220001 225000 5000 * 45 0 4.256839e-01 7.718511e-01
46 chr1 225001 230000 5000 * 46 0 4.256839e-01 7.718511e-01
47 chr1 230001 235000 5000 * 47 0 4.256839e-01 7.718511e-01
48 chr1 235001 240000 5000 * 48 0 4.256839e-01 7.718511e-01
49 chr1 240001 245000 5000 * 49 0 4.256839e-01 7.718511e-01
50 chr1 245001 250000 5000 * 50 0 4.256839e-01 7.718511e-01
51 chr1 250001 255000 5000 * 51 0 4.256839e-01 7.718511e-01
52 chr1 255001 260000 5000 * 52 0 4.256839e-01 7.718511e-01
53 chr1 260001 265000 5000 * 53 0 4.256839e-01 7.718511e-01
54 chr1 265001 270000 5000 * 54 0 4.256839e-01 7.718511e-01
55 chr1 270001 275000 5000 * 55 0 4.256839e-01 7.718511e-01
56 chr1 275001 280000 5000 * 56 0 4.256839e-01 7.718511e-01
57 chr1 280001 285000 5000 * 57 0 4.256839e-01 7.718511e-01
58 chr1 285001 290000 5000 * 58 1 1.071823e-01 2.066926e-01
59 chr1 290001 295000 5000 * 59 0 4.256839e-01 7.718511e-01
60 chr1 295001 300000 5000 * 60 0 4.256839e-01 7.718511e-01
61 chr1 300001 305000 5000 * 61 0 4.256839e-01 7.718511e-01
62 chr1 305001 310000 5000 * 62 0 4.256839e-01 7.718511e-01
63 chr1 310001 315000 5000 * 63 0 4.256839e-01 7.718511e-01
64 chr1 315001 320000 5000 * 64 0 4.256839e-01 7.718511e-01
65 chr1 320001 325000 5000 * 65 0 4.256839e-01 7.718511e-01
66 chr1 325001 330000 5000 * 66 0 4.256839e-01 7.718511e-01
67 chr1 330001 335000 5000 * 67 0 4.256839e-01 7.718511e-01
68 chr1 335001 340000 5000 * 68 0 4.256839e-01 7.718511e-01
69 chr1 340001 345000 5000 * 69 0 4.256839e-01 7.718511e-01
70 chr1 345001 350000 5000 * 70 0 4.256839e-01 7.718511e-01
71 chr1 350001 355000 5000 * 71 0 4.256839e-01 7.718511e-01
72 chr1 355001 360000 5000 * 72 0 4.256839e-01 7.718511e-01
73 chr1 360001 365000 5000 * 73 0 4.256839e-01 7.718511e-01
74 chr1 365001 370000 5000 * 74 0 4.256839e-01 7.718511e-01
75 chr1 370001 375000 5000 * 75 0 4.256839e-01 7.718511e-01
76 chr1 375001 380000 5000 * 76 0 4.256839e-01 7.718511e-01
77 chr1 380001 385000 5000 * 77 0 4.256839e-01 7.718511e-01
78 chr1 385001 390000 5000 * 78 0 4.256839e-01 7.718511e-01
79 chr1 390001 395000 5000 * 79 0 4.256839e-01 7.718511e-01
80 chr1 395001 400000 5000 * 80 0 4.256839e-01 7.718511e-01
81 chr1 400001 405000 5000 * 81 0 4.256839e-01 7.718511e-01
82 chr1 405001 410000 5000 * 82 0 4.256839e-01 7.718511e-01
83 chr1 410001 415000 5000 * 83 0 4.256839e-01 7.718511e-01
84 chr1 415001 420000 5000 * 84 0 4.256839e-01 7.718511e-01
85 chr1 420001 425000 5000 * 85 0 4.256839e-01 7.718511e-01
86 chr1 425001 430000 5000 * 86 0 4.256839e-01 7.718511e-01
87 chr1 430001 435000 5000 * 87 0 4.256839e-01 7.718511e-01
88 chr1 435001 440000 5000 * 88 0 4.256839e-01 7.718511e-01
89 chr1 440001 445000 5000 * 89 0 4.256839e-01 7.718511e-01
90 chr1 445001 450000 5000 * 90 0 4.256839e-01 7.718511e-01
binomialPvalue nlog_pval
1 1.000000e+00 0.0000000
2 1.000000e+00 0.0000000
3 4.271127e-01 0.8507073
4 1.000000e+00 0.0000000
5 1.000000e+00 0.0000000
6 1.000000e+00 0.0000000
7 1.000000e+00 0.0000000
8 1.000000e+00 0.0000000
9 1.000000e+00 0.0000000
10 1.000000e+00 0.0000000
11 1.000000e+00 0.0000000
12 1.000000e+00 0.0000000
13 1.065385e-01 2.2392486
14 1.000000e+00 0.0000000
15 1.000000e+00 0.0000000
16 4.271127e-01 0.8507073
17 1.000000e+00 0.0000000
18 1.000000e+00 0.0000000
19 1.000000e+00 0.0000000
20 1.000000e+00 0.0000000
21 1.000000e+00 0.0000000
22 1.000000e+00 0.0000000
23 1.000000e+00 0.0000000
24 1.000000e+00 0.0000000
25 1.000000e+00 0.0000000
26 1.000000e+00 0.0000000
27 1.000000e+00 0.0000000
28 1.000000e+00 0.0000000
29 1.000000e+00 0.0000000
30 1.000000e+00 0.0000000
31 1.000000e+00 0.0000000
32 1.000000e+00 0.0000000
33 1.000000e+00 0.0000000
34 1.000000e+00 0.0000000
35 1.000000e+00 0.0000000
36 1.000000e+00 0.0000000
37 1.000000e+00 0.0000000
38 1.000000e+00 0.0000000
39 1.000000e+00 0.0000000
40 1.000000e+00 0.0000000
41 1.000000e+00 0.0000000
42 1.000000e+00 0.0000000
43 1.000000e+00 0.0000000
44 1.000000e+00 0.0000000
45 1.000000e+00 0.0000000
46 1.000000e+00 0.0000000
47 1.000000e+00 0.0000000
48 1.000000e+00 0.0000000
49 1.000000e+00 0.0000000
50 1.000000e+00 0.0000000
51 1.000000e+00 0.0000000
52 1.000000e+00 0.0000000
53 1.000000e+00 0.0000000
54 1.000000e+00 0.0000000
55 1.000000e+00 0.0000000
56 1.000000e+00 0.0000000
57 1.000000e+00 0.0000000
58 4.271127e-01 0.8507073
59 1.000000e+00 0.0000000
60 1.000000e+00 0.0000000
61 1.000000e+00 0.0000000
62 1.000000e+00 0.0000000
63 1.000000e+00 0.0000000
64 1.000000e+00 0.0000000
65 1.000000e+00 0.0000000
66 1.000000e+00 0.0000000
67 1.000000e+00 0.0000000
68 1.000000e+00 0.0000000
69 1.000000e+00 0.0000000
70 1.000000e+00 0.0000000
71 1.000000e+00 0.0000000
72 1.000000e+00 0.0000000
73 1.000000e+00 0.0000000
74 1.000000e+00 0.0000000
75 1.000000e+00 0.0000000
76 1.000000e+00 0.0000000
77 1.000000e+00 0.0000000
78 1.000000e+00 0.0000000
79 1.000000e+00 0.0000000
80 1.000000e+00 0.0000000
81 1.000000e+00 0.0000000
82 1.000000e+00 0.0000000
83 1.000000e+00 0.0000000
84 1.000000e+00 0.0000000
85 1.000000e+00 0.0000000
86 1.000000e+00 0.0000000
87 1.000000e+00 0.0000000
88 1.000000e+00 0.0000000
89 1.000000e+00 0.0000000
90 1.000000e+00 0.0000000
[ reached getOption("max.print") -- omitted 605623 rows ]
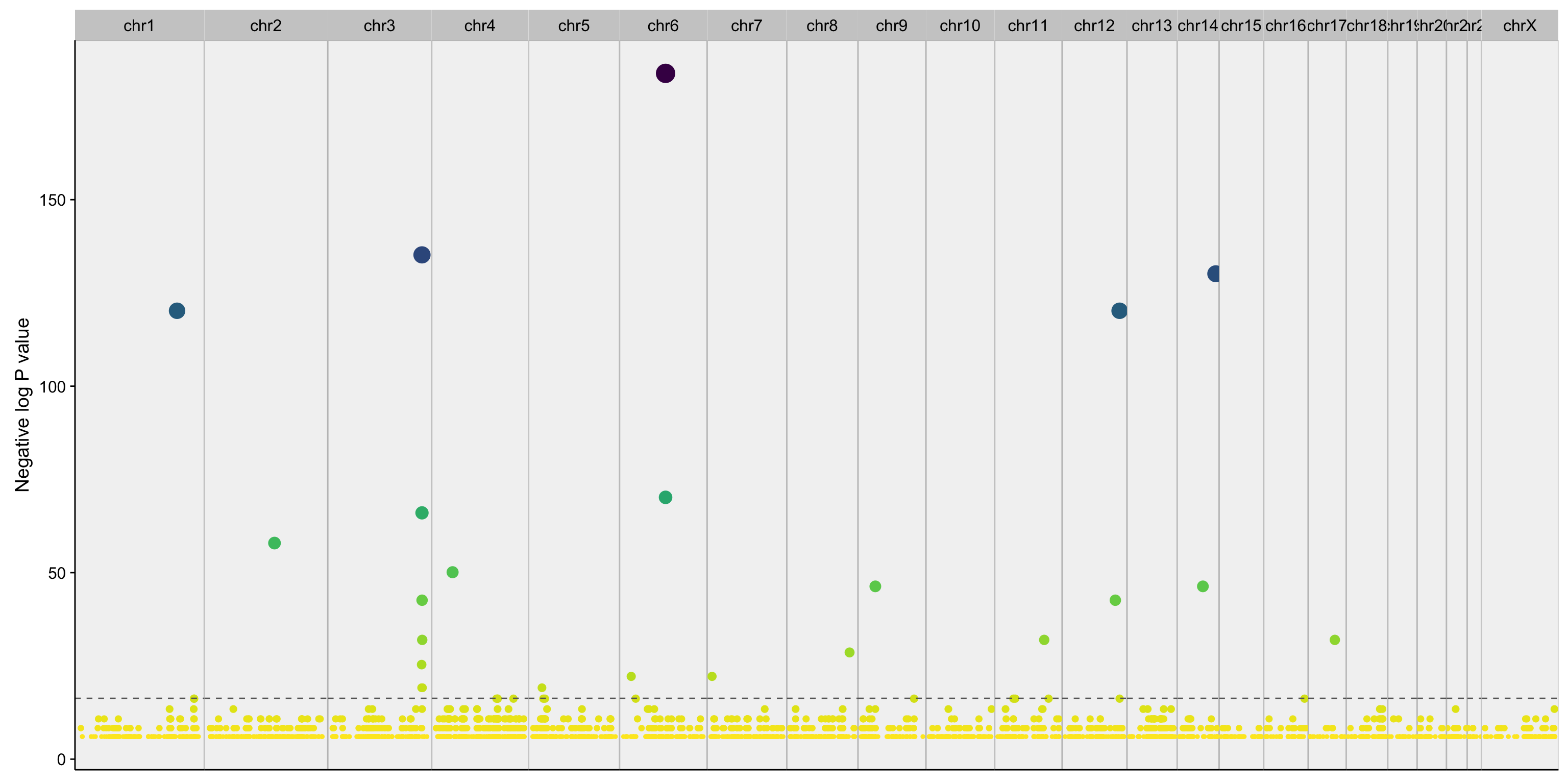
manhattanplot_maf_noncoding_mut_density <-
mut_counts_df %>%
filter(nlog_pval > 5) %>%
ggplot(aes(start, nlog_pval, color = nlog_pval, size = nlog_pval)) +
geom_point() +
geom_hline(yintercept = -log(0.05/nrow(mut_counts_df)), linetype = 2, colour = "grey40") +
scale_x_continuous(labels = NULL) +
viridis::scale_color_viridis(breaks = NULL, direction = -1) +
scale_size_continuous(breaks = NULL) +
facet_grid(~seqnames, scales = "free_x", space = "free_x") +
theme(
panel.background = element_rect(fill = "grey95", linetype = 1, size = 0.5, colour = "grey75"),
panel.spacing = unit(0, "lines"),
axis.ticks.x = element_blank()) +
labs(x = NULL, y = "Negative log P value")
manhattanplot_maf_noncoding_mut_density
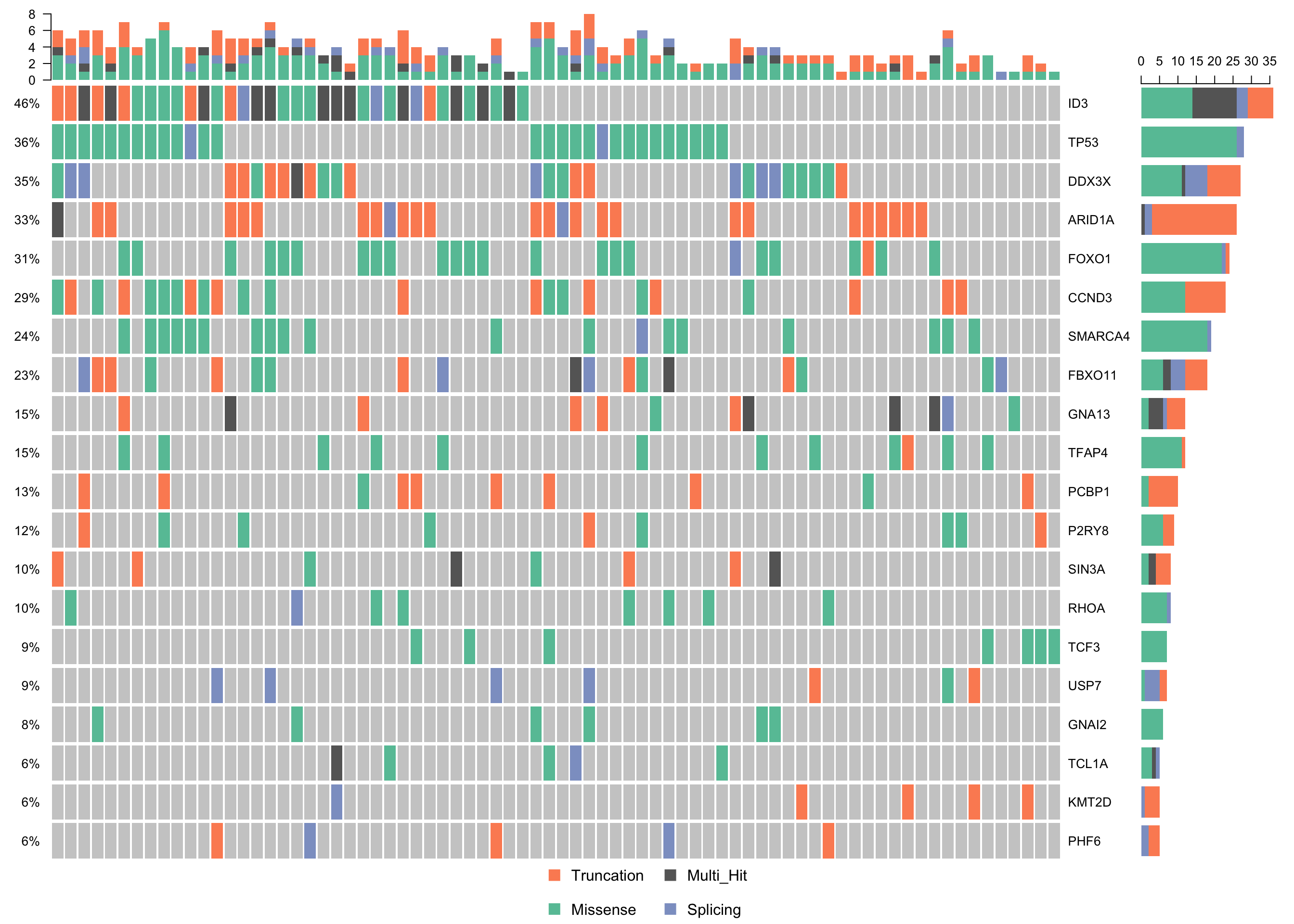
waterfallplot_mmaf <-
subsetMaf(mmaf, query = "Variant_Classification %in% c('Truncation', 'Missense', 'Splicing')",
mafObj = TRUE) %>%
oncoplot(genes = smgs, colors = colours$categs, )
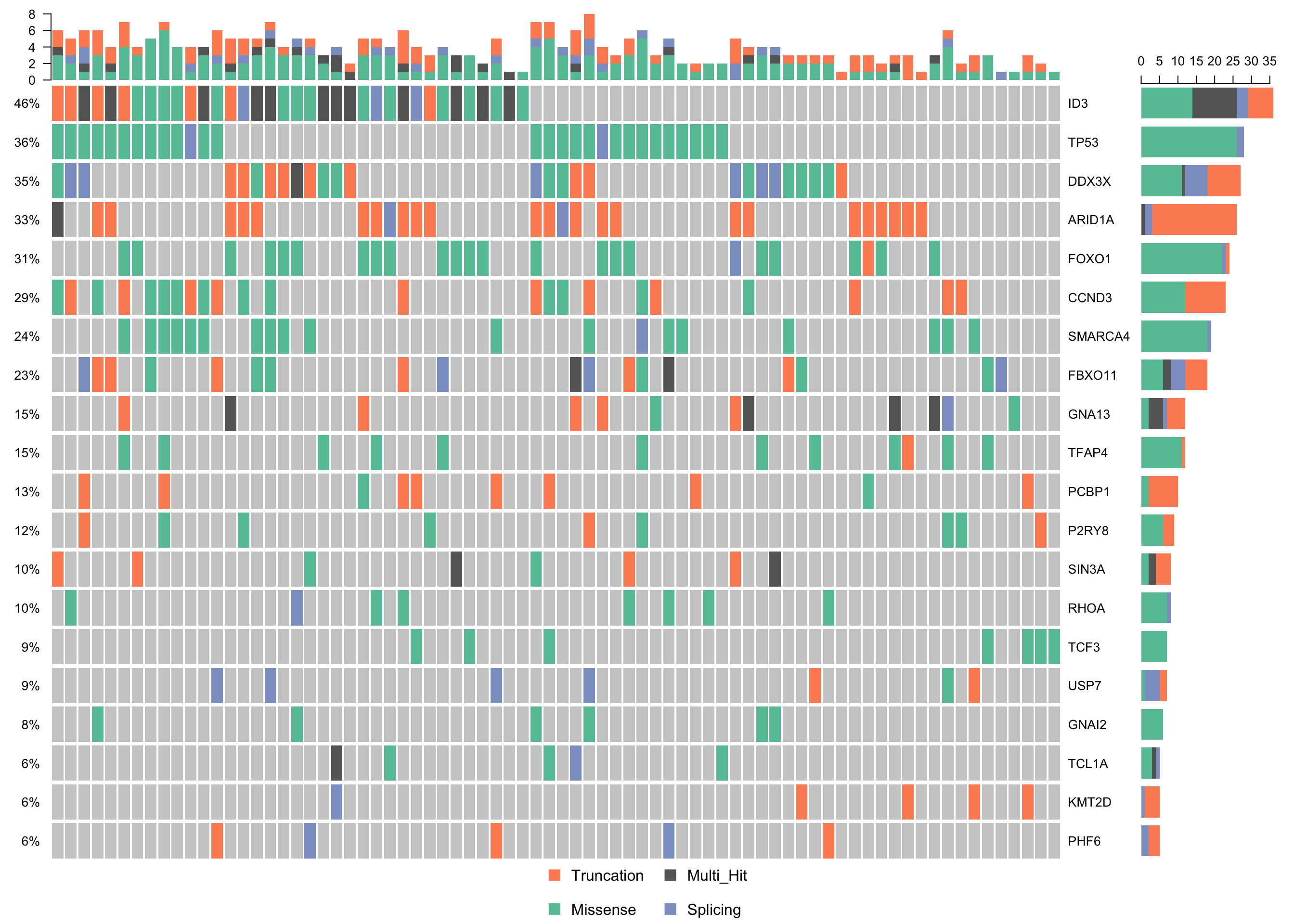
print(waterfallplot_mmaf)
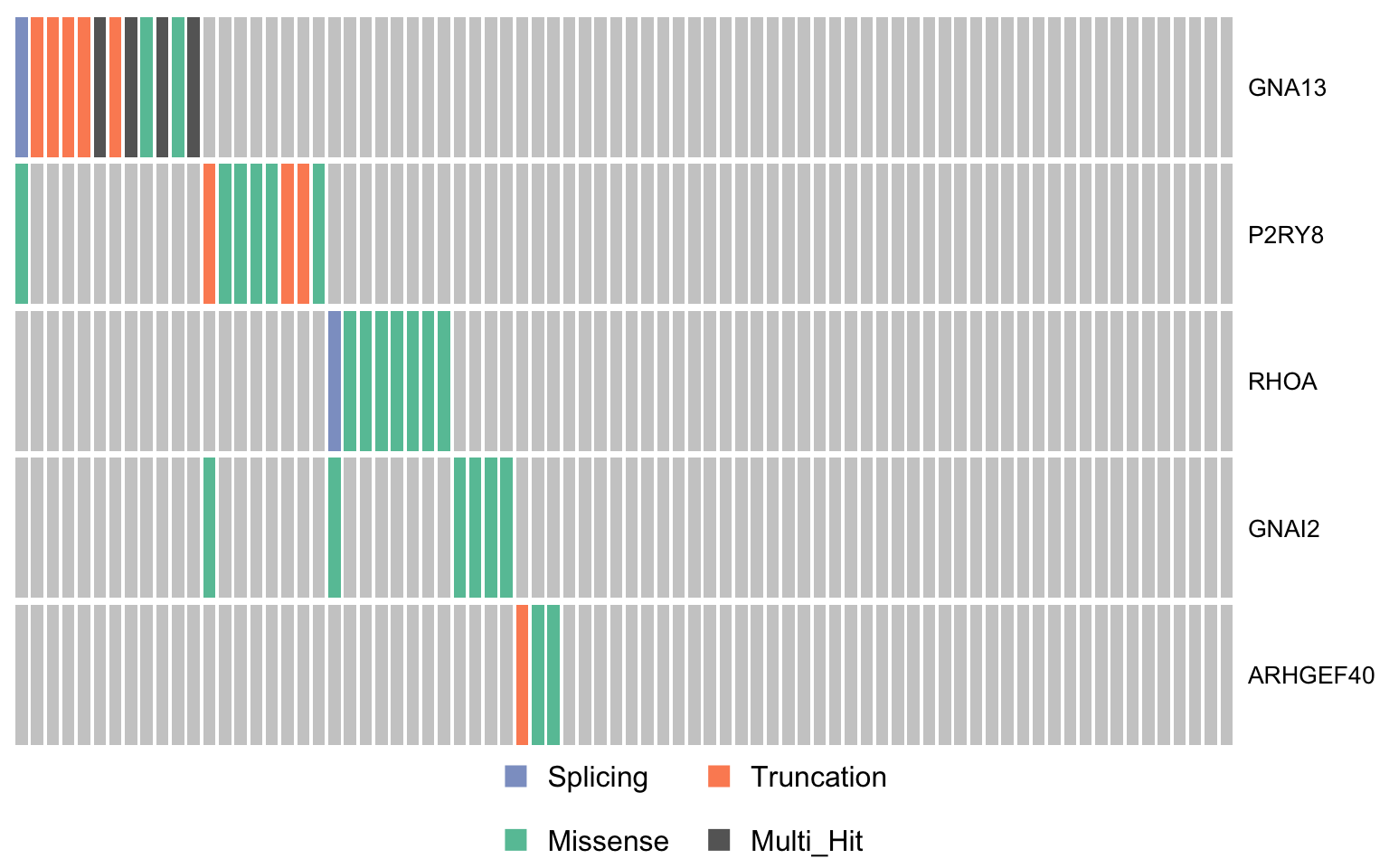
gprotein_genes <- c("GNA13", "P2RY8", "RHOA", "GNAI2", "ARHGEF40")
# mmaf@data %>%
# filter(Hugo_Symbol %in% gprotein_genes) %>%
# select(Hugo_Symbol, Tumor_Sample_Barcode) %>%
# bind_rows(tibble(Hugo_Symbol = "DROP", Tumor_Sample_Barcode = unique(mmaf@data$Tumor_Sample_Barcode))) %>%
# mutate(status = 0) %>%
# distinct() %>%
# spread(Hugo_Symbol, status, fill = 1, drop = FALSE) %>%
# select(-Tumor_Sample_Barcode, -DROP) %>%
# map(as.factor) %>%
# table() %>%
# c() %>%
# cometExactTest::comet_exact_test()
oncostrip_maf_gproteins <- oncostrip(mmaf, genes = gprotein_genes,
colors = colours$categs, removeNonMutated = FALSE)
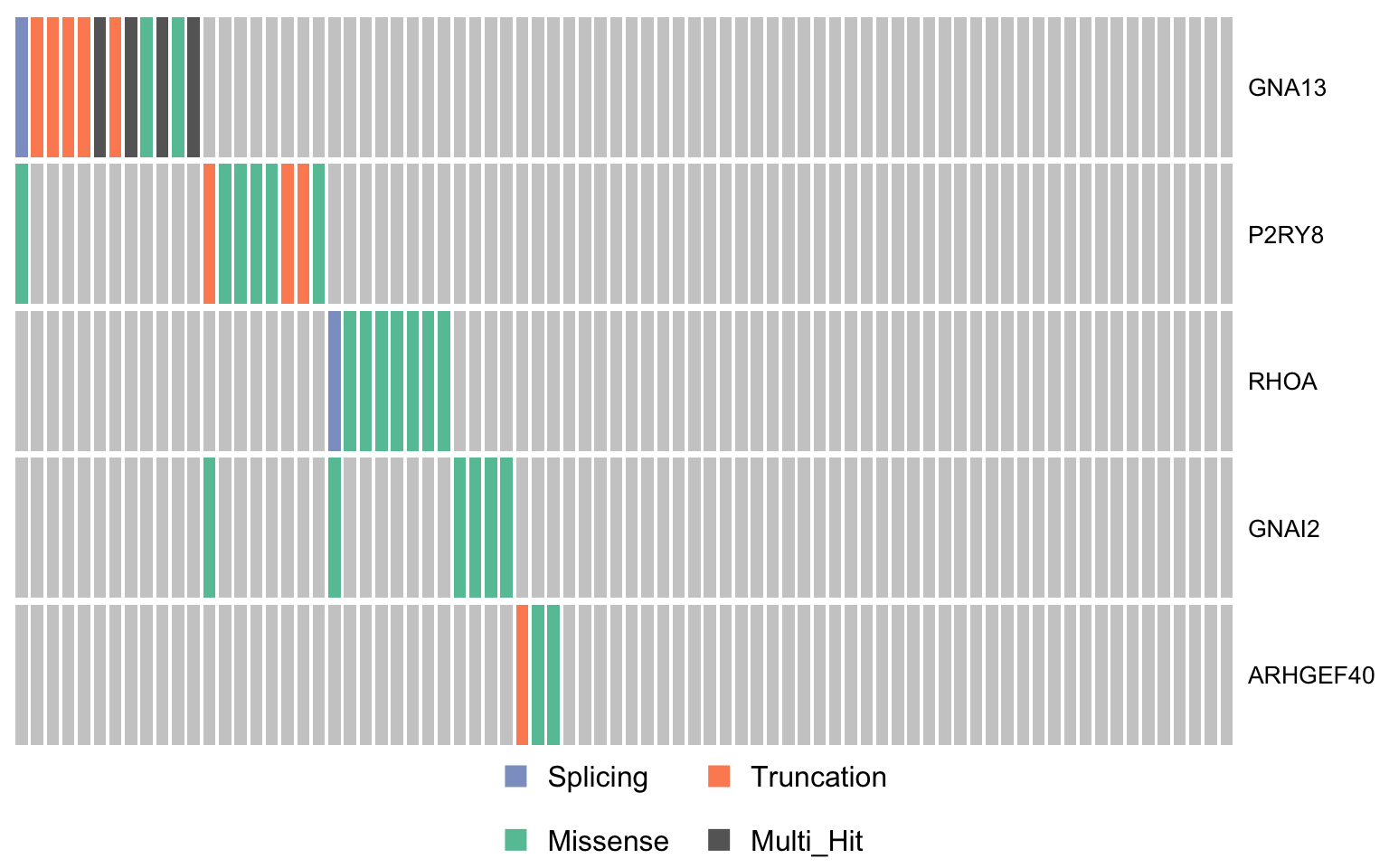
print(oncostrip_maf_gproteins)
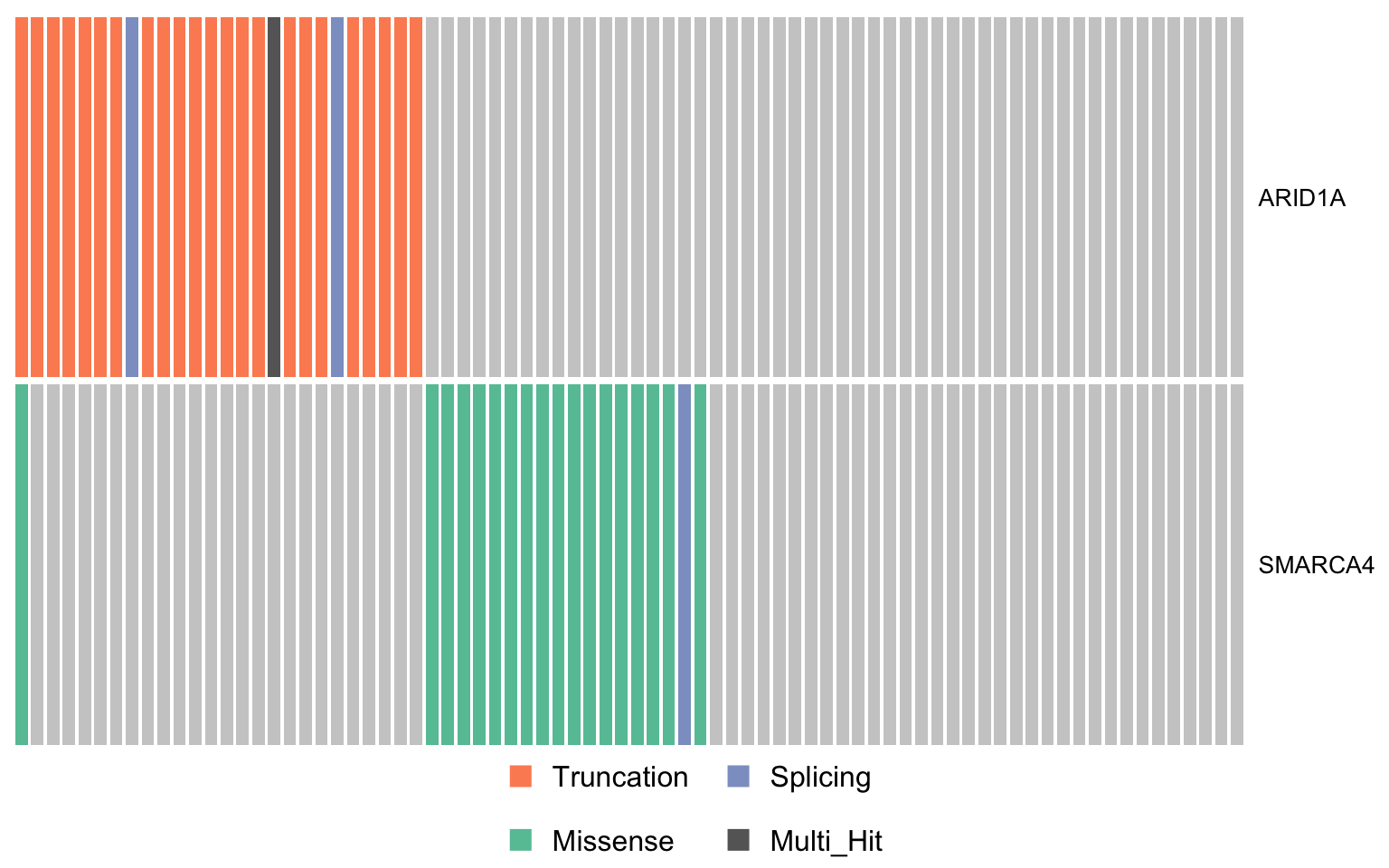
swisnf_genes <- c("ARID1A", "SMARCA4")
oncostrip_maf_swisnf <- oncostrip(
mmaf, genes = swisnf_genes, colors = colours$categs, removeNonMutated = FALSE)
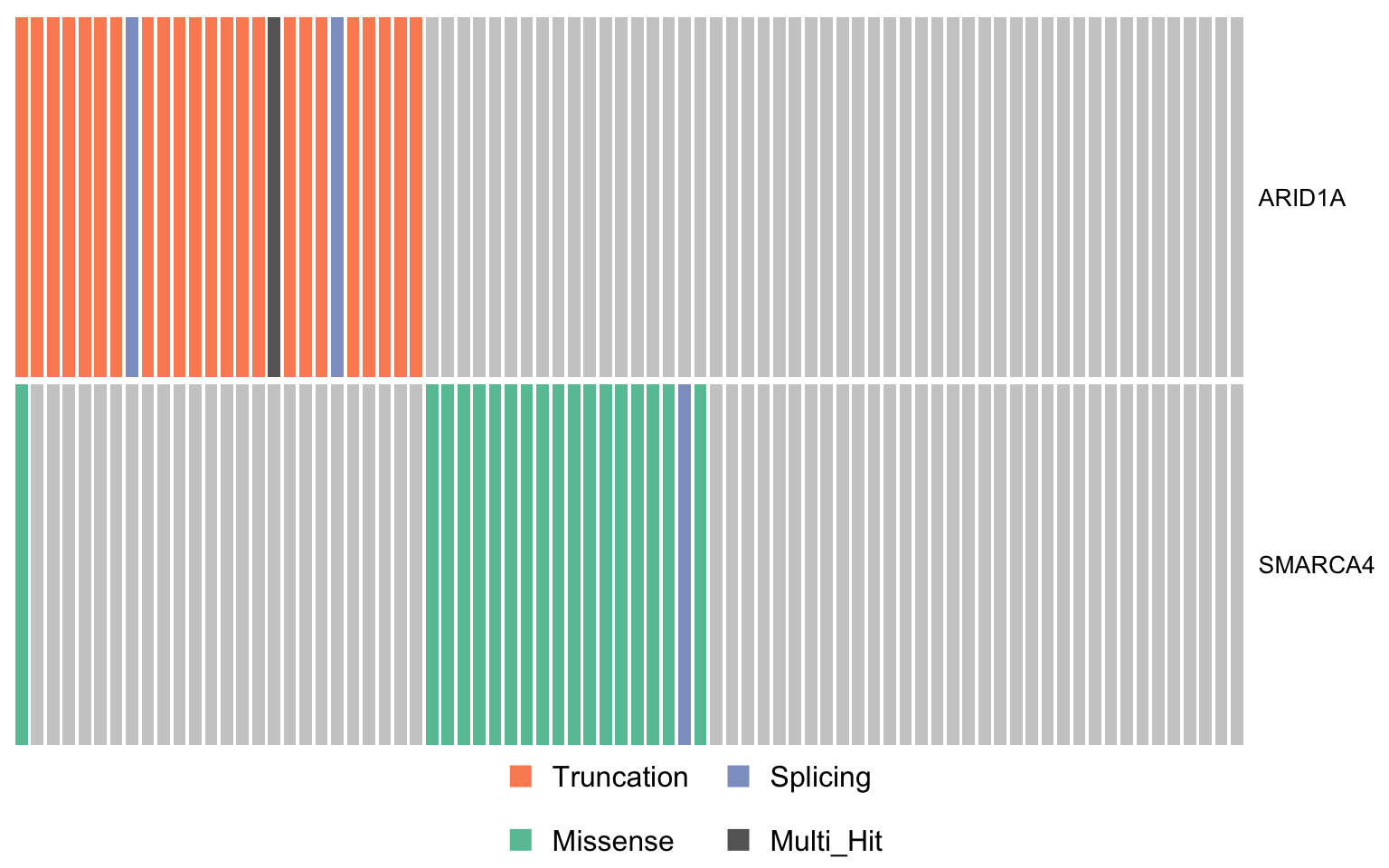
print(oncostrip_maf_swisnf)