HVNC1
Bruno Grande
2017-02-18 14:31:03
HVCN1-mutated Cases per COO
hvcn1 <-
muts %>%
dplyr::filter(gene == "HVCN1")
patients_hvcn1 <- unique(hvcn1$patient)
coo_hvcn1 <-
coo %>%
dplyr::filter(patient %in% muts$patient,
coo_ns %in% c("ABC", "GCB")) %>%
dplyr::mutate(hvcn1_status = patient %in% patients_hvcn1)
coo_hvcn1 %>%
dplyr::group_by(coo_ns) %>%
dplyr::summarize(hvcn1_mutant = sum(hvcn1_status),
total = length(patient),
prop = round(hvcn1_mutant / total, 3)) %>%
knitr::kable()| coo_ns | hvcn1_mutant | total | prop |
|---|---|---|---|
| ABC | 1 | 95 | 0.011 |
| GCB | 7 | 170 | 0.041 |
Fisher’s Exact Test
Testing for the association of HVCN1 mutations with COO.
fisher_hvcn1 <-
coo_hvcn1 %>%
dplyr::mutate(hvcn1_status = patient %in% patients_hvcn1) %$%
table(coo_ns, hvcn1_status) %>%
fisher.test()
fisher_hvcn1
Fisher's Exact Test for Count Data
data: .
p-value = 0.2656
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
0.504025 183.694360
sample estimates:
odds ratio
4.020788 Differential Expression of HVCN1
Boxplots
Across all DLBCL samples
texpr_hvcn1_all <-
texpr["HVCN1", , drop = FALSE] %>%
t() %>%
as.data.frame() %>%
tibble::rownames_to_column("patient") %>%
dplyr::mutate(hvcn1_status = ifelse(patient %in% patients_hvcn1, "Mutated", "Not Mutated")) %>%
dplyr::rename(expr = HVCN1)
boxplot_hvcn1_expr_all <-
texpr_hvcn1_all %>%
ggplot(aes(x = hvcn1_status, y = expr)) +
geom_boxplot() +
geom_jitter(position = position_jitter(width = 0.5, height = 0))
boxplot_hvcn1_expr_all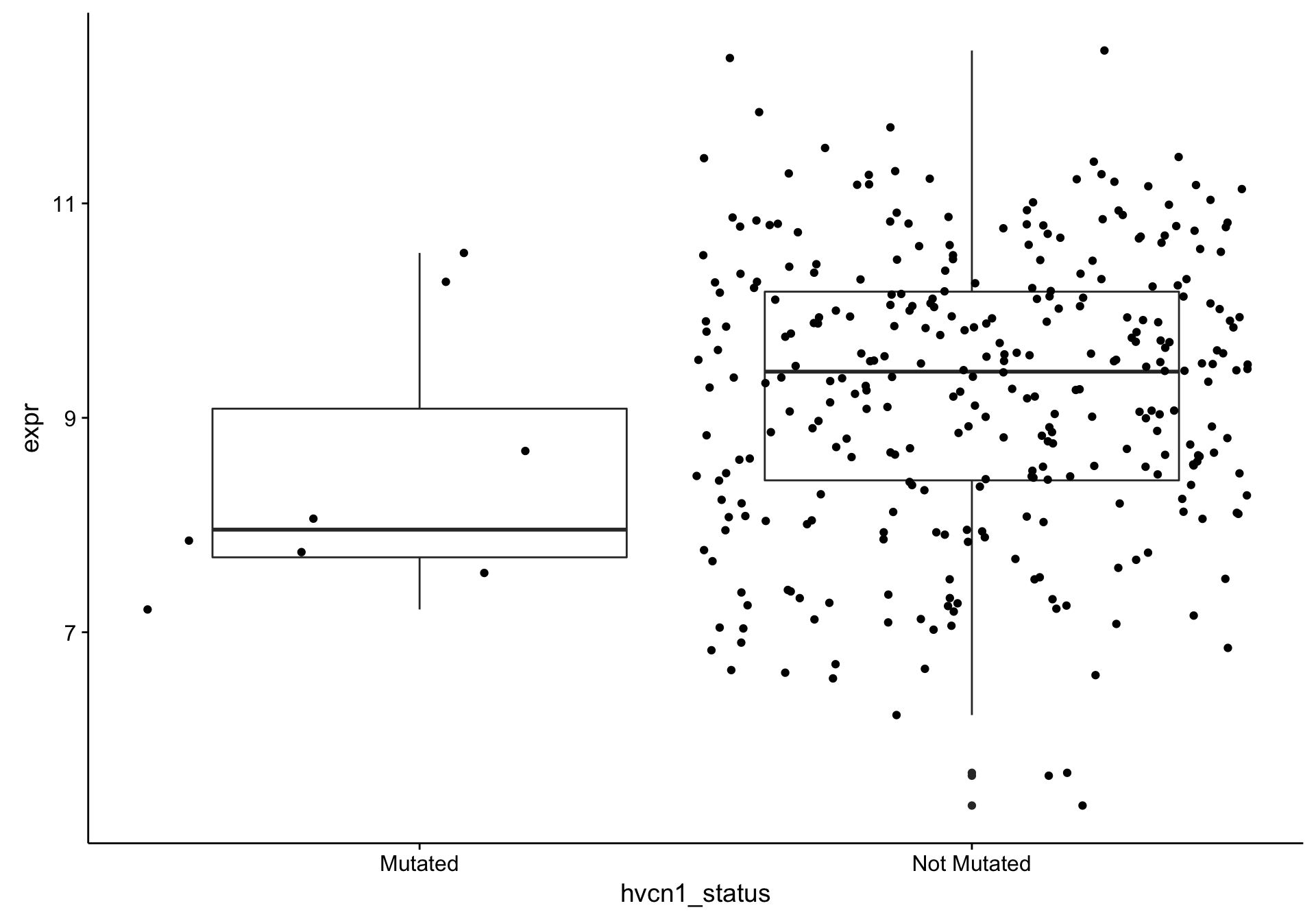
Across GCB DLBCL samples
texpr_hvcn1_gcb <-
texpr_hvcn1_all %>%
dplyr::left_join(coo, by = "patient") %>%
dplyr::filter(coo_ns == "GCB")
boxplot_hvcn1_expr_gcb <-
boxplot_hvcn1_expr_all %+% texpr_hvcn1_gcb
boxplot_hvcn1_expr_gcb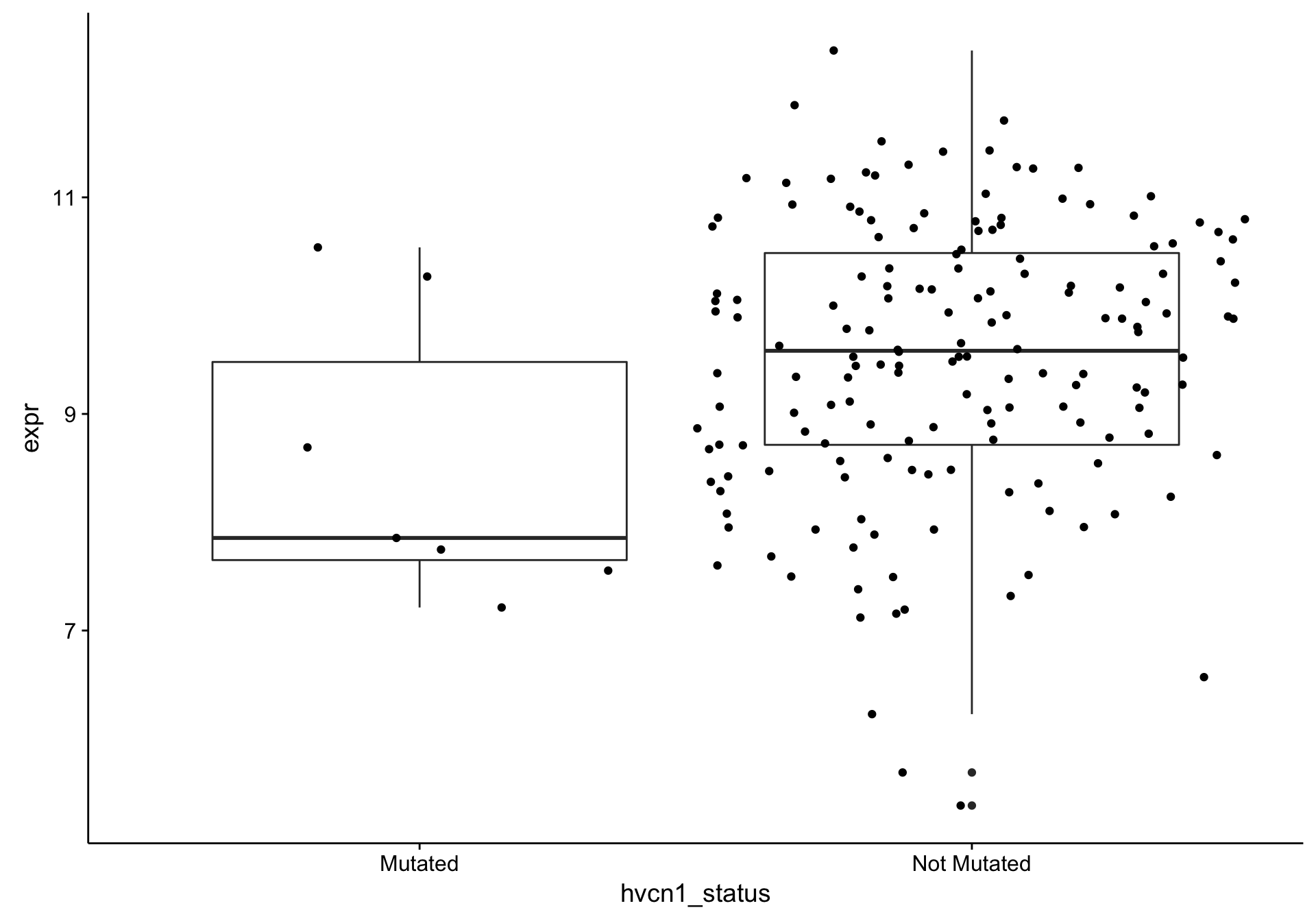
Mann-Whitney U test
Using all DLBCL samples
hvcn1_mutants <- texpr_hvcn1_all$patient %in% patients_hvcn1
wilcox.test(texpr_hvcn1_gcb[hvcn1_mutants, "expr"], texpr_hvcn1_gcb[!hvcn1_mutants, "expr"])
Wilcoxon rank sum test with continuity correction
data: texpr_hvcn1_gcb[hvcn1_mutants, "expr"] and texpr_hvcn1_gcb[!hvcn1_mutants, "expr"]
W = 575, p-value = 0.01398
alternative hypothesis: true location shift is not equal to 0Using GCB DLBCL samples
hvcn1_mutants <- texpr_hvcn1_gcb$patient %in% patients_hvcn1
wilcox.test(texpr_hvcn1_gcb[hvcn1_mutants, "expr"], texpr_hvcn1_gcb[!hvcn1_mutants, "expr"])
Wilcoxon rank sum test with continuity correction
data: texpr_hvcn1_gcb[hvcn1_mutants, "expr"] and texpr_hvcn1_gcb[!hvcn1_mutants, "expr"]
W = 327, p-value = 0.05465
alternative hypothesis: true location shift is not equal to 0