FASSI is a stand-alone Perl application that calculates BAC end and BAC overlap length constraints from clone fingerprint map contigs created by the FPC package to improve the contiguity of whole-genome shotgun sequence (WGS) assemblies. The FASSI method is simple to implement, potentially cost-effective and has resulted in the increase of large scaffold contiguity and coverage for both the Drosophila melanogaster and Cryptococcus gattii genomes. A 6.5X draft DNA sequence of the Pan troglodytes (Chimpanzee) genome was assembled using map-derived constraints and resulted in a 26.1% increase in scaffold contiguity.
Summary
FASSI is written in PERL and runs on Linux. The software produces three types of length constraints, supplied to PCAP (Huang et al. 2003) as a permissible lower and upper size limit between any two BAC end sequences having a relationship. Relationships include BAC ends from the same clone, logical intrinsic and logical reverse-complemented BAC ends between overlapping or buried clones. Although the output of FASSI is currently formatted to work with the PCAP assembly program, it can be easily formatted to work with other assembly algorithms able the use length constraints for individual clones.
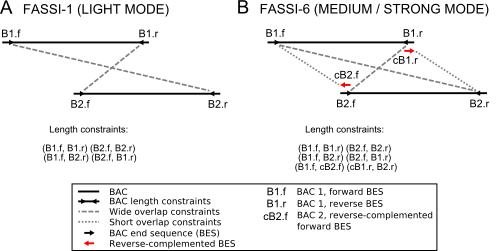
BAC length constraints derived from physical maps. (A) FASSI-1 (FASSI-LIGHT) consists of four different sets of length constraints between any two overlapping BAC neighbors (40% overlap in our implementation). In addition to introducing accurate length constraints between mate pairs of a given BAC, two additional, wide overlap constraints are introduced between neighbors. (B) Constraints in FASSI-6 (MEDIUM MODE) include the length constraints from FASSI-1, plus short overlap constraints between any two proximal BAC end sequences from BAC neighbours sharing 40% of their length. FASSI ran in STRONG mode will generate length, overlap and buried constraints between any overlapping BACs (not just neighbours).
License
Copyright (c) 2006-2020 Canada's Michael Smith Genome Science Centre. All rights reserved.
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
Results
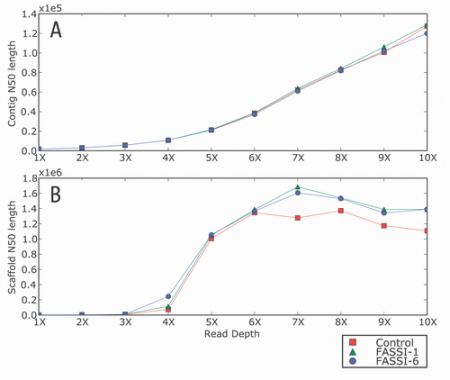
Effect of FASSI length constraints on the assembly contiguity of Drosophila melanogaster. (A) Contig and (B) scaffold N50 length were calculated for every assembly at shotgun depths ranging from 1X to 10X.
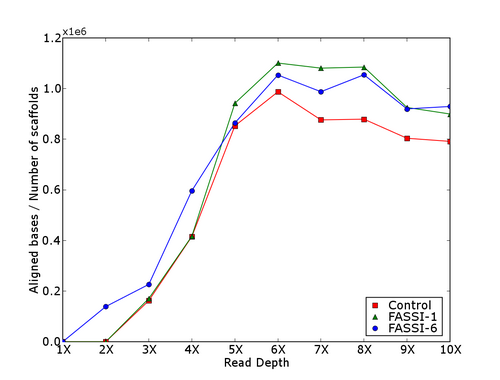
Contiguity Score for Drosophila assemblies having > hundred million aligned bases to the finished genome. This analysis shows that the genome is covered by fewer and larger scaffolds.
Documentation
Warren RL, Varabei D, Platt D, Huang X, Messina D, Yang S-P, Kronstad JW, Krzywinski M, Warren WC, Wallis JW, Hillier LW, Chinwalla AT, Schein JE, Siddiqui AS, Marra MA, Wilson RK, Jones SJM. 2006. Physical map-assisted whole-genome shotgun sequence assemblies. Genome Res. 16:768-775
Credits
British Columbia Cancer Agency - Genome Sciences Centre
René Warren, Dmitry Varabei, Martin Krzywinski, Jacqueline Schein, Asim Siddiqui, Marco Marra and Steven Jones
Washington University School of Medicine - Genome Sequencing Center
David Messina, Shiaw-Pyng Yang, John Wallis, LaDeanna Hillier, Asif Chinwalla, and Richard Wilson
Department of Computer Science - Iowa State University
Xiaoqiu Huang
Joint Genome Institute
Darren Platt
Funding for this work was provided by a grant from the National Human Genome Research Institute (NHGRI).
Current Release
FASSI - Fingerprint and Assembly Incorporation 1.0
Released Jul 20, 2006
All Releases
| Version | Released | Description | Licenses | Status |
|---|---|---|---|---|
| 1.0 | Jul 20, 2006 | First release | GPL | final |