 Using only a blood sample from a cancer patient, this project will validate Parent-of-Origin-Aware genomic analysis (POAga) in common, high penetrant hereditary cancer conditions such as hereditary breast and ovarian cancer (HBOC) and Lynch syndrome, rarer syndromes with parent-of-origin-effects and other genes predisposing to breast cancer and gastrointestinal malignancies that are associated with genes across multiple chromosomes.
Using only a blood sample from a cancer patient, this project will validate Parent-of-Origin-Aware genomic analysis (POAga) in common, high penetrant hereditary cancer conditions such as hereditary breast and ovarian cancer (HBOC) and Lynch syndrome, rarer syndromes with parent-of-origin-effects and other genes predisposing to breast cancer and gastrointestinal malignancies that are associated with genes across multiple chromosomes.
The Parent of Origin Aware Genomic Analysis (POAga) is a collaborative initiative lead by Dr. Intan Schrader from the Hereditary Cancer Program, Dr. Steve Jones from Genome Sciences Centre and Dr. Peter Lansdorp from BC Cancer Research Centre. The initiative is funded by multiple agencies including Canadian Institutes of Health Research (CIHR), Genome Canada and Genome BC.
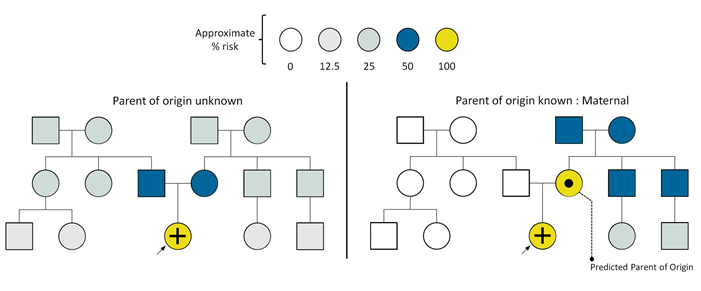
The Parent of Origin Aware Genomic Analysis combines nanopore sequencing and strand-seq technologies. Strand-seq provides chromosome-scale but sparse phasing of single nucleotide variants (SNVs). Phased SNVs from strand-seq are then passed to nanopore long-reads to provide chromosome-scale phasing of nearly all the genomic variants. Chromosome-scale haplotyping will result in two haplotypes (H1 and H2) each representing one parent. To assign parent-of-origin to H1 and H2, the informatician will then use DNA methylation data, which can be obtained from nanopore data, at known human imprinted regions. As we already know which allele is methylated (maternal or paternal) in these imprinted regions, we can assign a maternal or paternal origin to chromosome-scale haplotypes and genomic variants. This technology allows the assignment of parental segregation without parental data.
In this initiative, POAga will be performed using blood derived from patients with Pancreatic Cancer, Hereditary Breast and Ovarian Cancer, Lynch syndrome, hereditary paraganglioma-pheochromocytoma syndrome and CDH1-related Diffuse Gastric and Lobular Breast Cancer Syndrome (DGLBCS).