EXPRESSION ANALYSIS
This section provides details of the expression analyses performed as part of the POG pipeline. Look in this section for details about expression comparator choice and potentially relevance genes determined to be significant expression outliers.
EXPRESSION ANALYSIS
SPEARNMAN CORRELATION WITH AVAILABLE EXPRESSION DATASETS
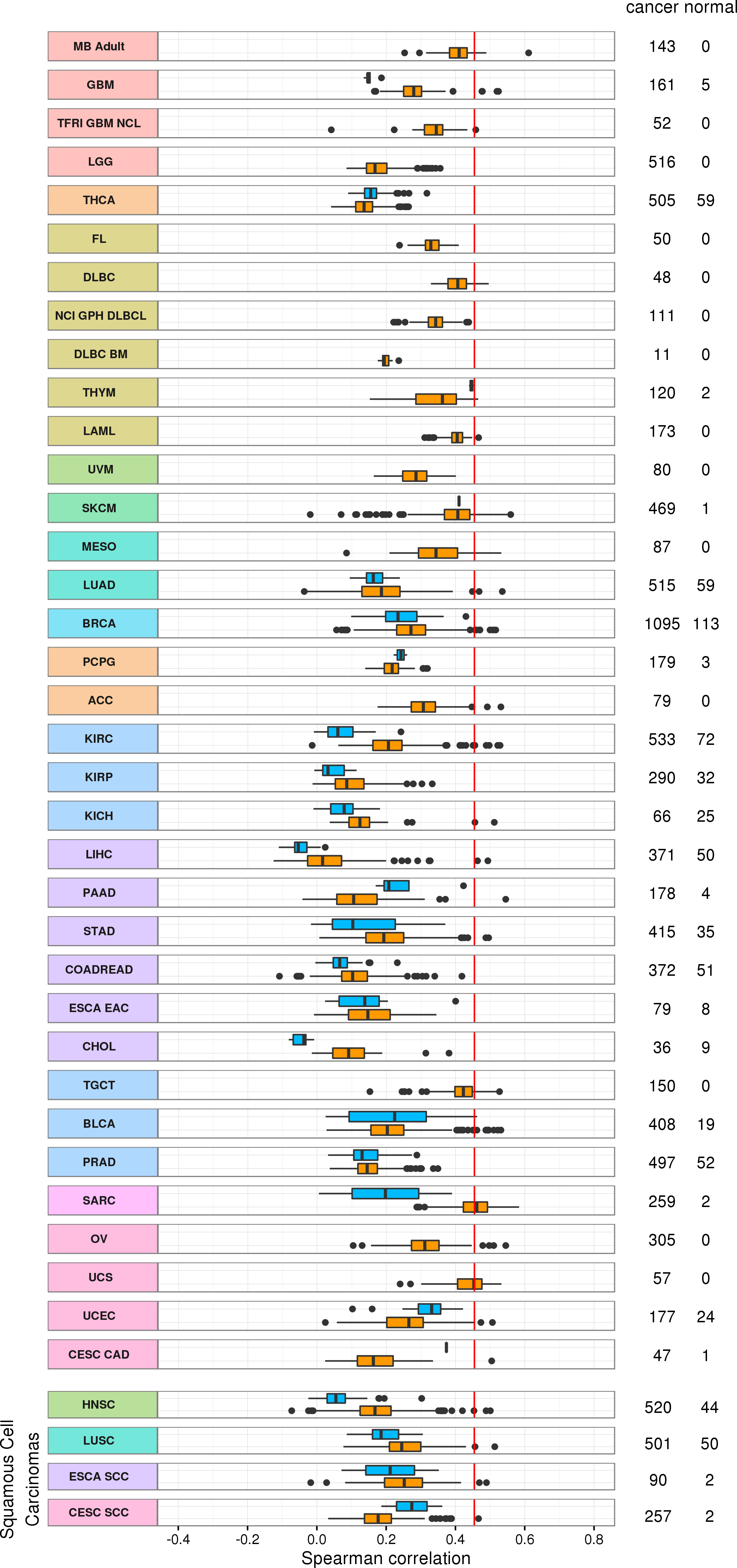 |
Normal Expression Comparator
Disease Expression Comparator
compendium average
SARC
|
EXPRESION LEVEL OUTLIERS OF POTENTIAL CLINICAL RELEVANCE
| Gene | Location | Copy Change | LOH State | CNV State | Expression (RPKM) | Fold Change vs. average |
SARC %ile |
|---|---|---|---|---|---|---|---|
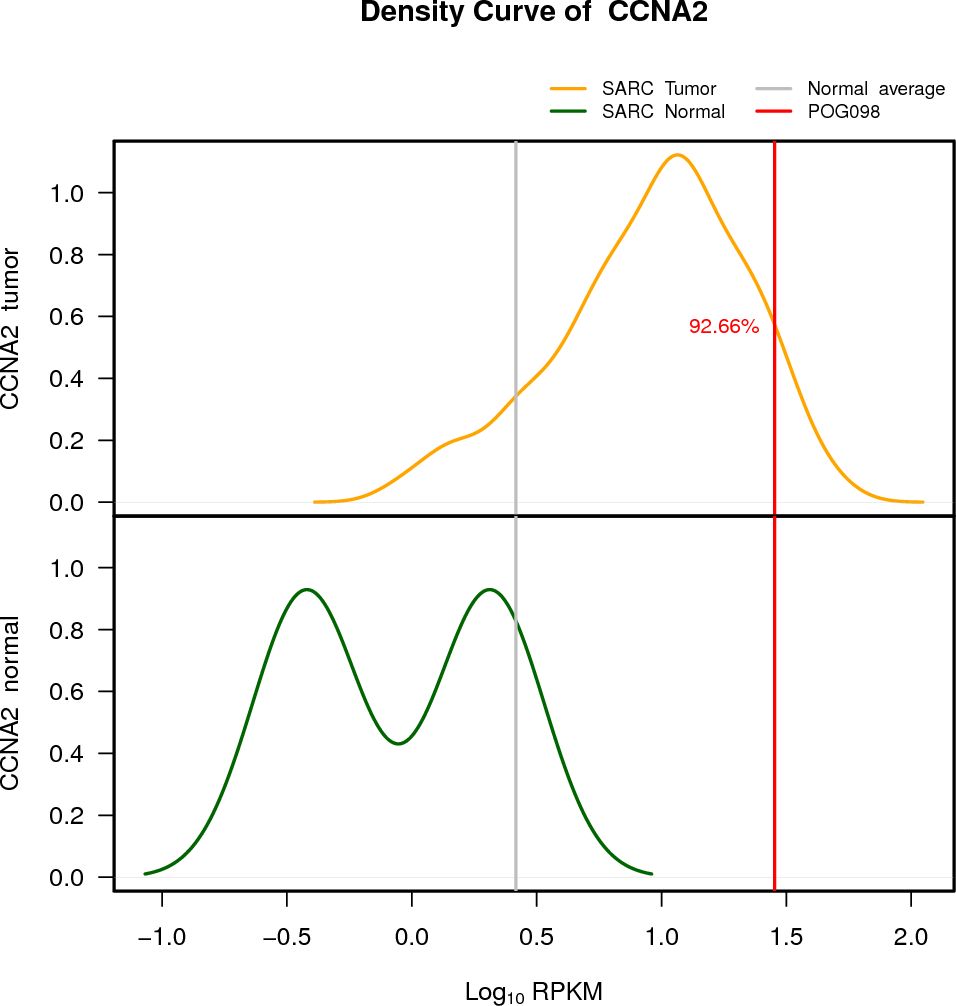
| CCNA2 | 4:122737599-122745087 | 0 | HET | Neutral | 28.38 | 8.13 | 93 |
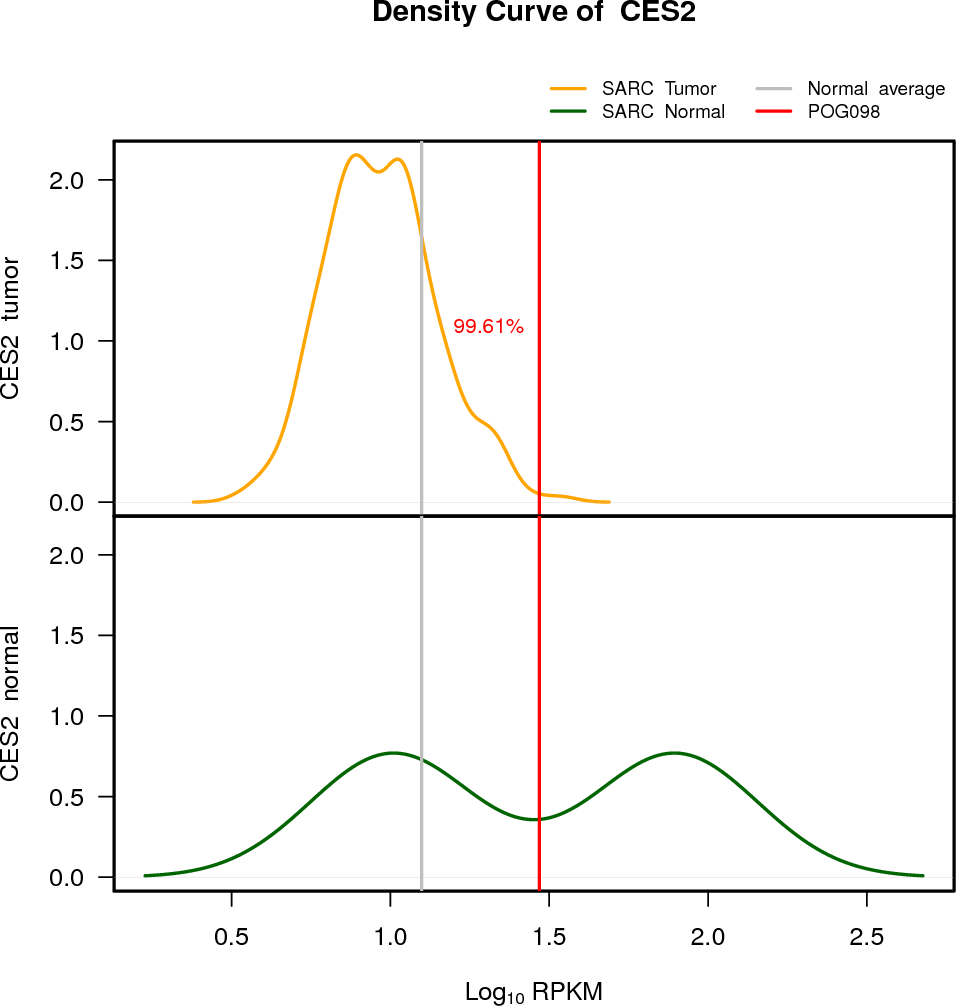
| CES2 | 16:66968347-66978999 | 0 | NLOH | Neutral | 29.43 | 2.25 | 100 |
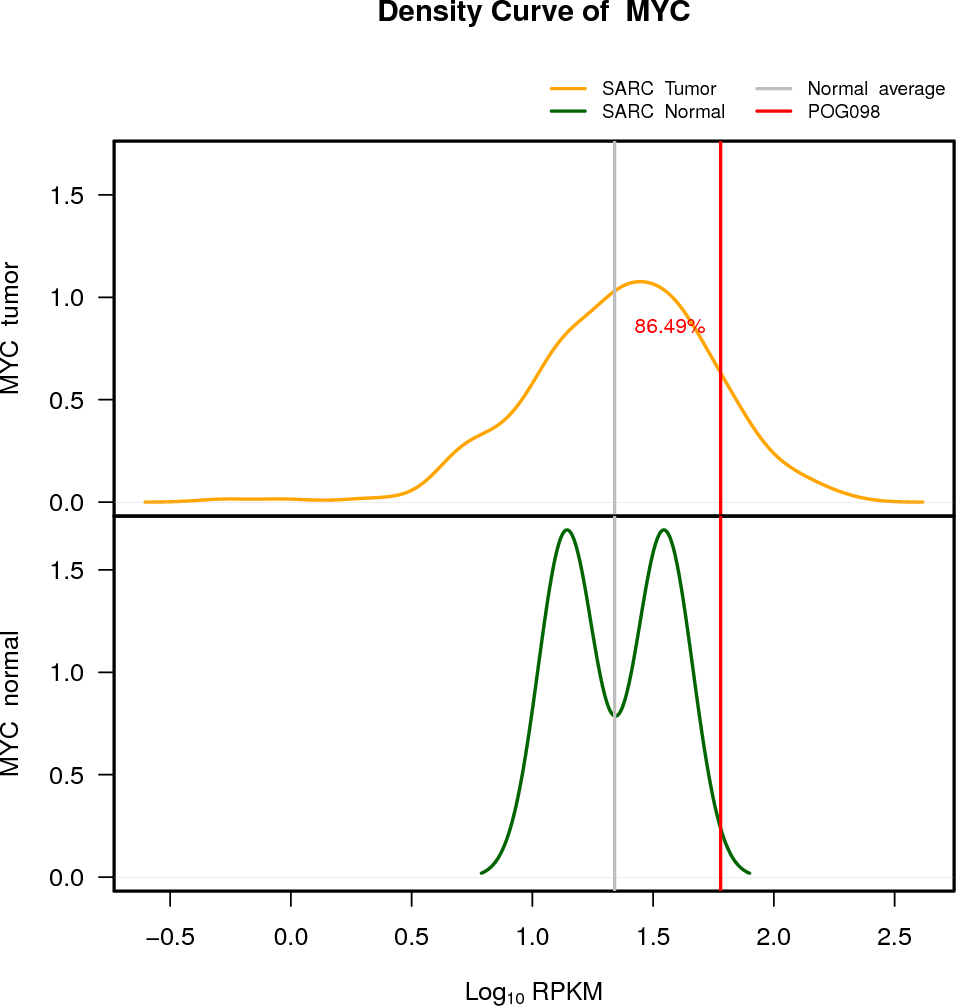
| MYC | 8:128747680-128753674 | +2 | ALOH | Gain | 60.13 | 2.67 | 86 |
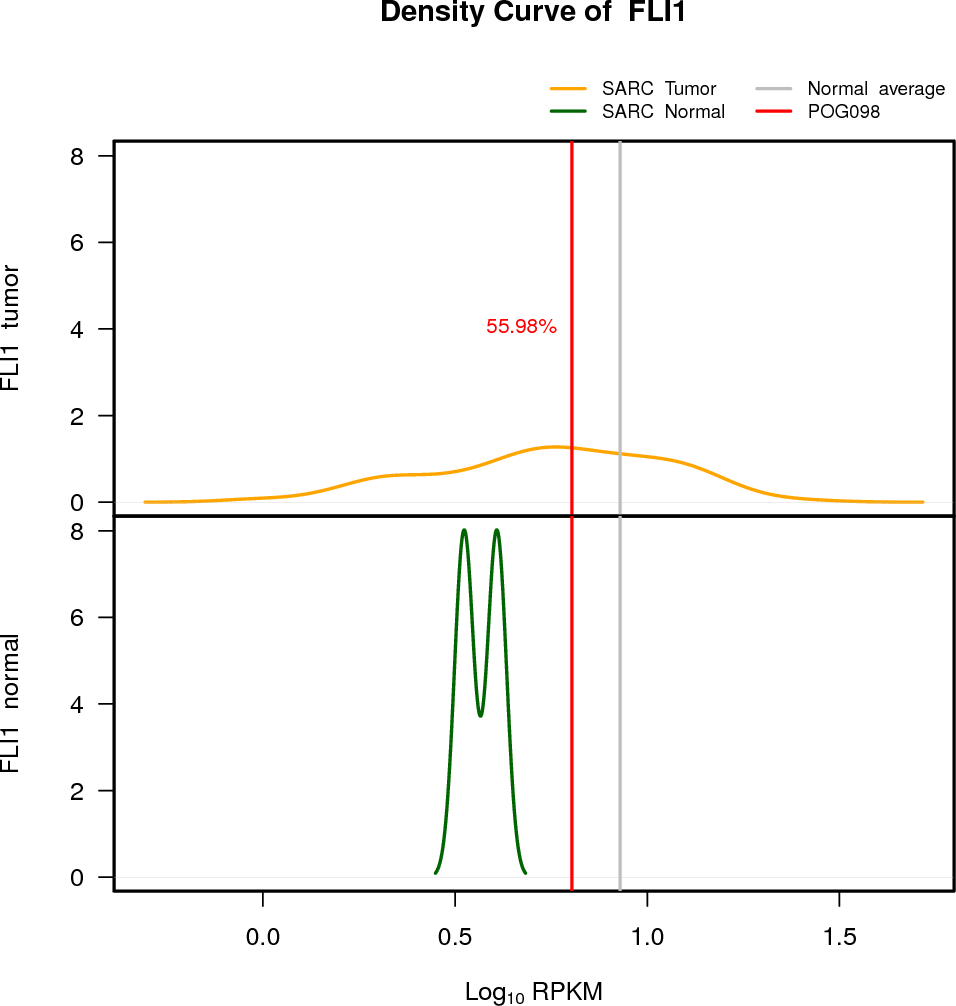
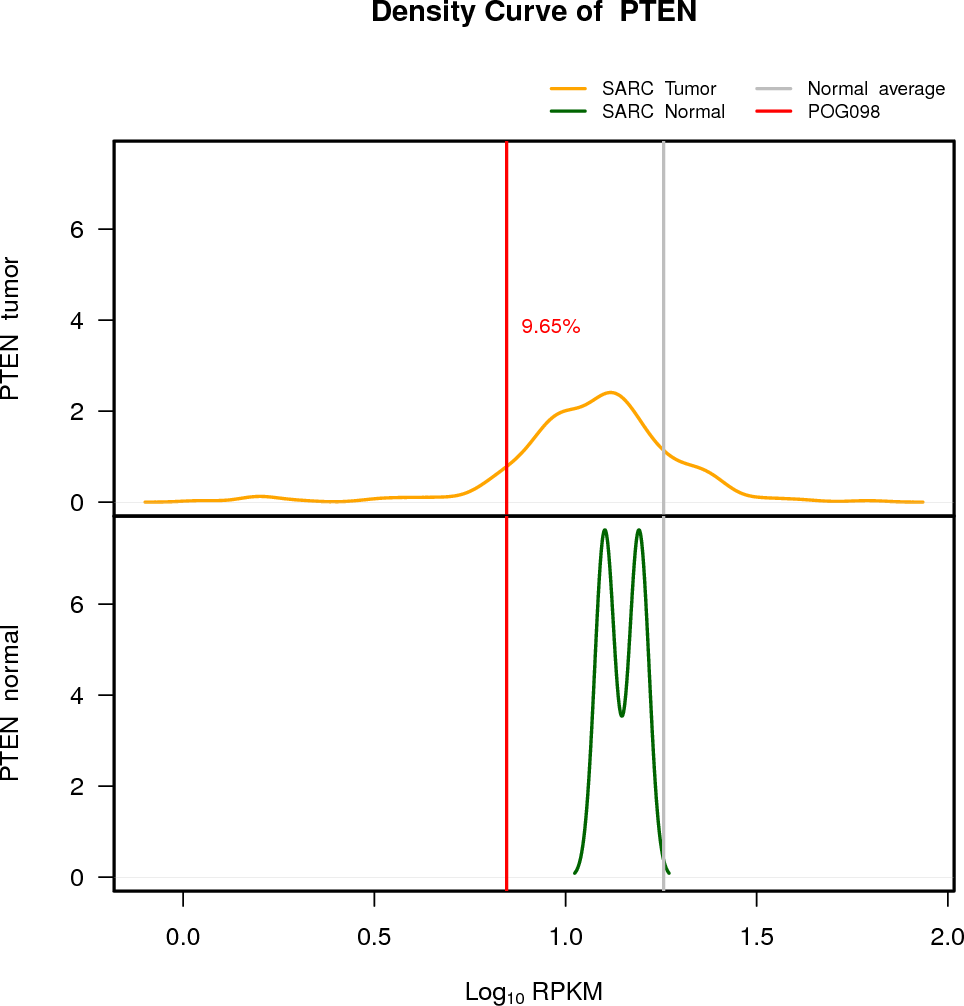
| PTEN | 10:89622870-89731687 | 0 | HET | Neutral | 7.02 | -2.38 | 10 |
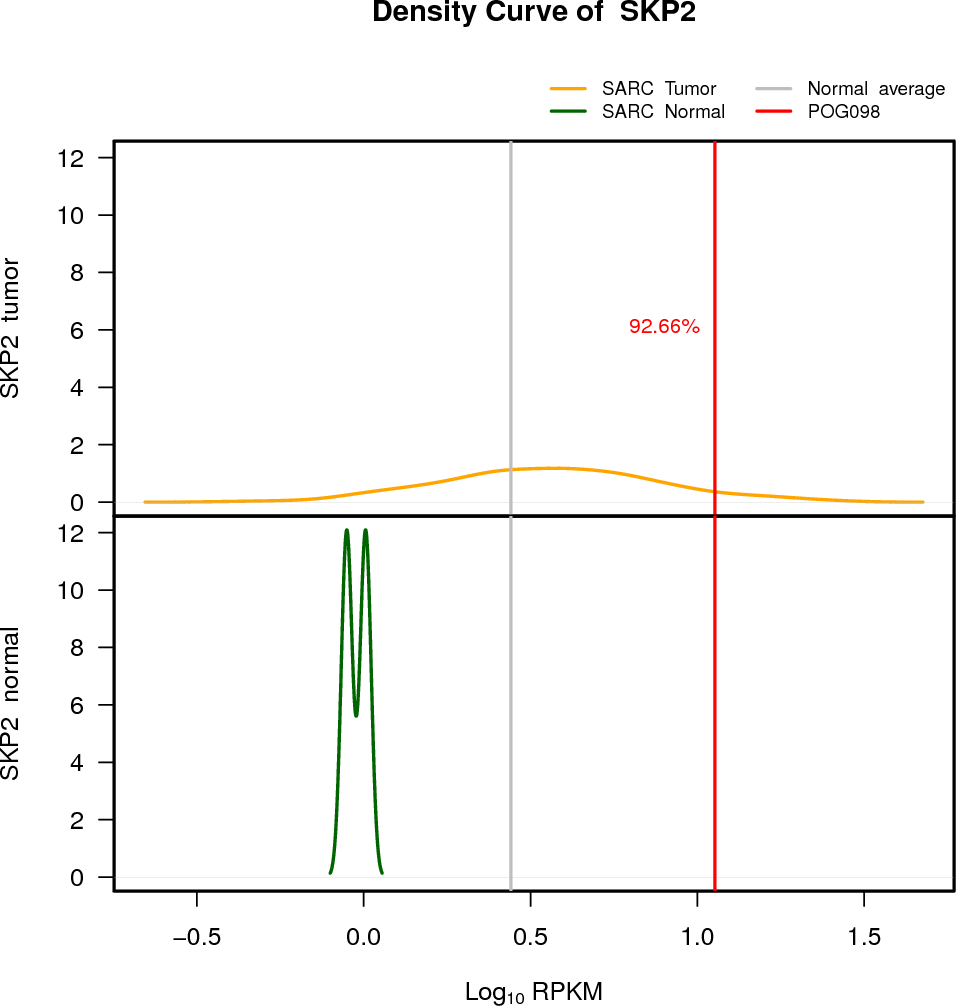
| SKP2 | 5:36152091-36184421 | 0 | HET | Neutral | 11.29 | 3.27 | 93 |
EXPRESION LEVEL OUTLIERS OF PROGNOSTIC AND DIAGNOSTIC RELEVANCE
| Gene | Location | Copy Change | LOH State | CNV State | Expression (RPKM) | Fold Change vs. average |
SARC %ile |
|---|---|---|---|---|---|---|---|
| CES2 | 16:66968347-66978999 | 0 | NLOH | Neutral | 29.43 | 2.25 | 100 |
EXPRESION LEVEL OUTLIERS OF BIOLOGICAL RELEVANCE
| Gene | Location | Copy Change | LOH State | CNV State | Expression (RPKM) | Fold Change vs. average |
SARC %ile |
|---|---|---|---|---|---|---|---|
| CCNA2 | 4:122737599-122745087 | 0 | HET | Neutral | 28.38 | 8.13 | 93 |
| CES2 | 16:66968347-66978999 | 0 | NLOH | Neutral | 29.43 | 2.25 | 100 |
| MYC | 8:128747680-128753674 | +2 | ALOH | Gain | 60.13 | 2.67 | 86 |
| PTEN | 10:89622870-89731687 | 0 | HET | Neutral | 7.02 | -2.38 | 10 |
| SKP2 | 5:36152091-36184421 | 0 | HET | Neutral | 11.29 | 3.27 | 93 |
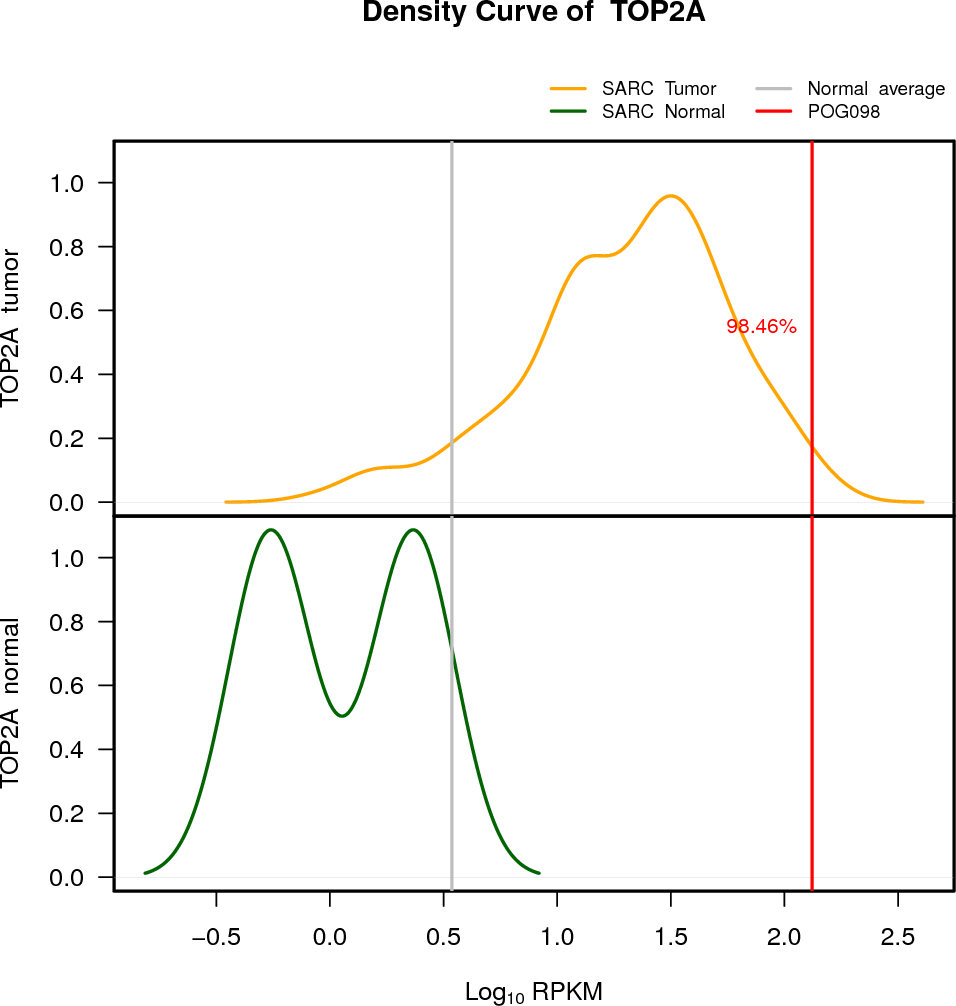
| TOP2A | 17:38544768-38574202 | 0 | HET | Neutral | 132.57 | 30.1 | 98 |
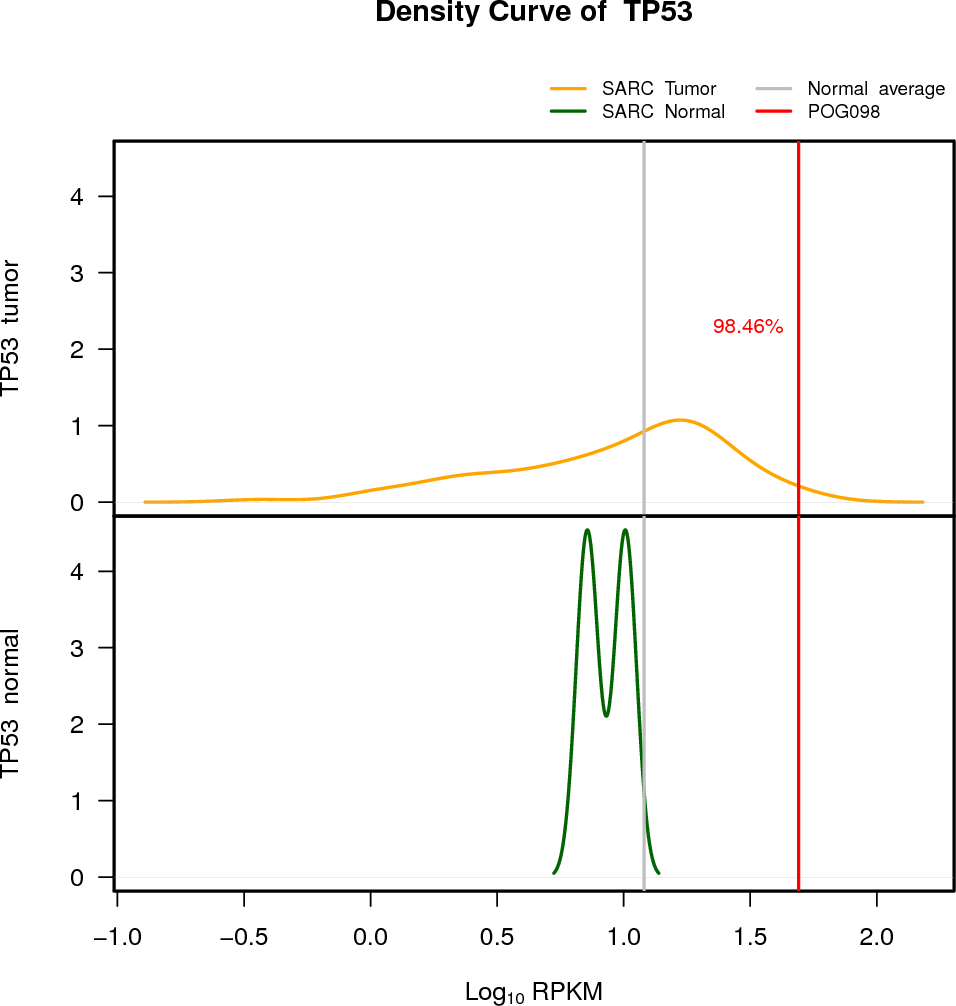
| TP53 | 17:7565097-7590856 | 0 | HET | Neutral | 49.13 | 3.85 | 98 |
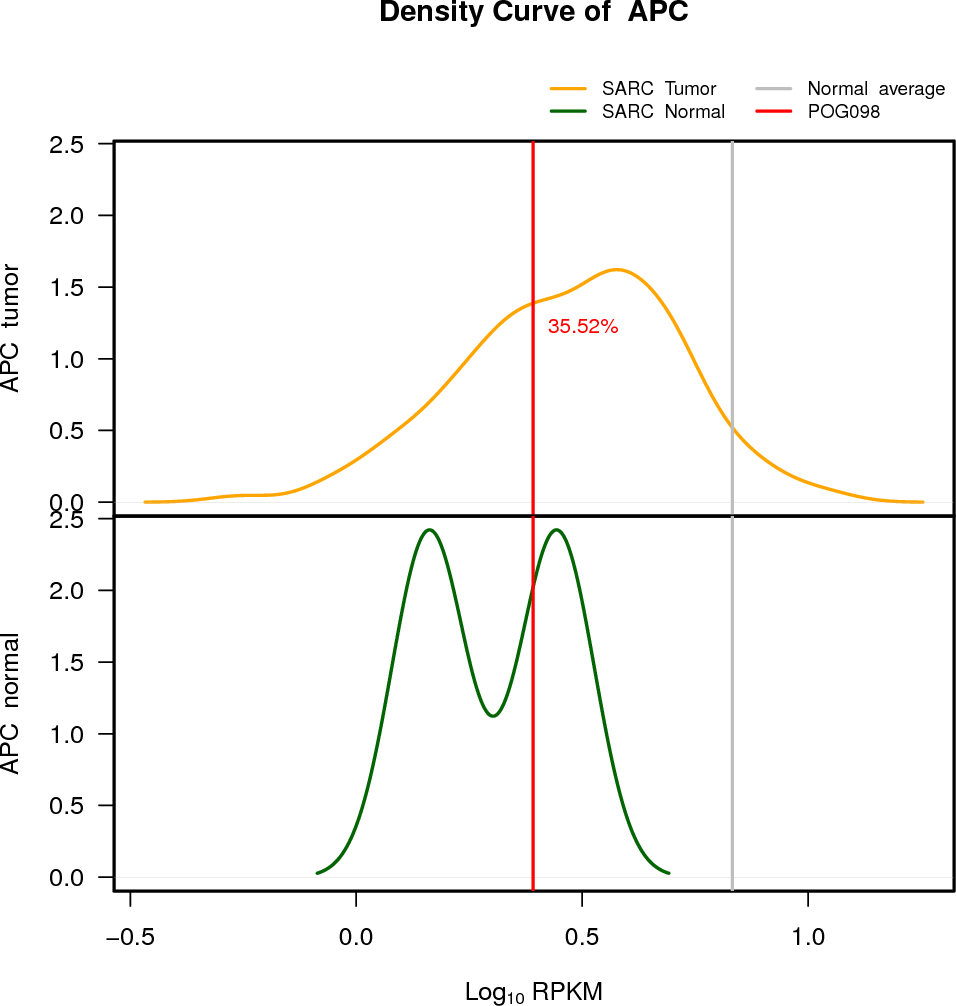
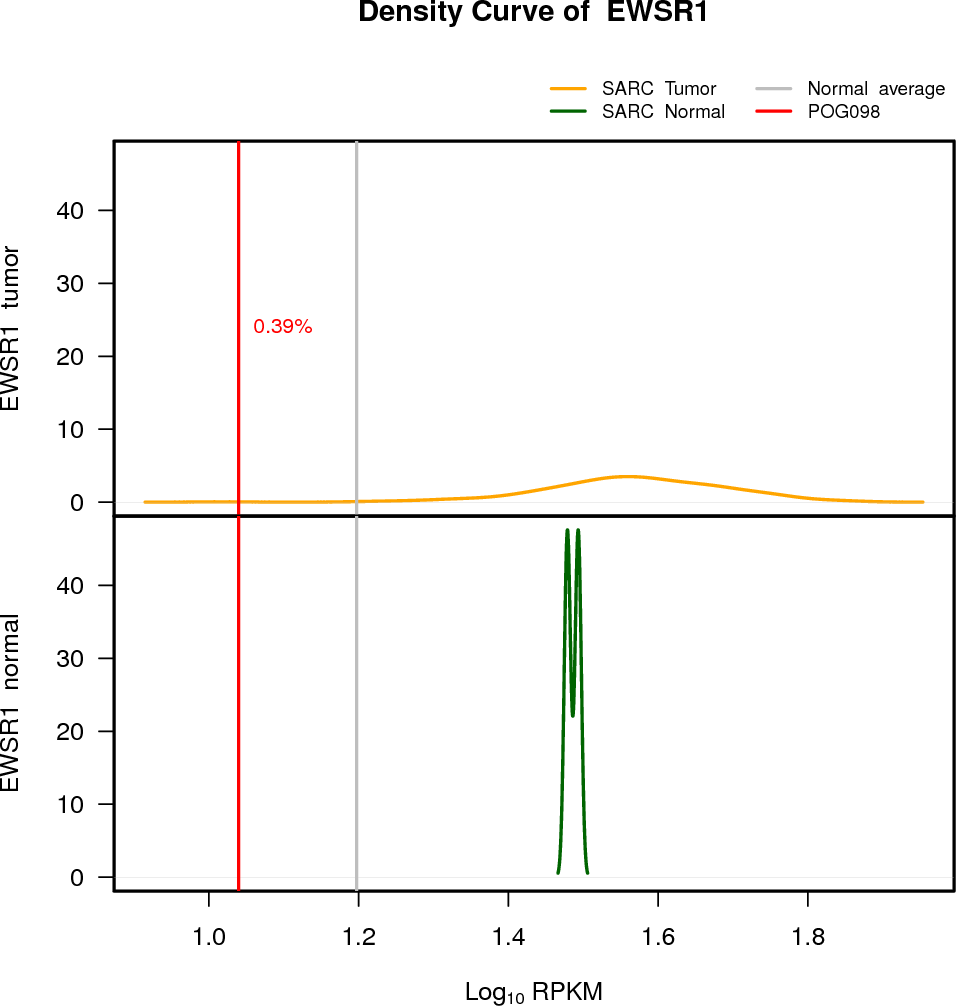
EXPRESION DISTRIBUTIONS