COPY NUMBER ANALYSIS
This section provides details of copy number alterations detected in the tumour, including gains and losses and loss of heterozygosity (LOH). Look in this section for details about potentially relevant genes within the copy change regions and their associated expression metrics.
COPY NUMBER AND LOH
SUMMARY OF COPY NUMBER EVENTS
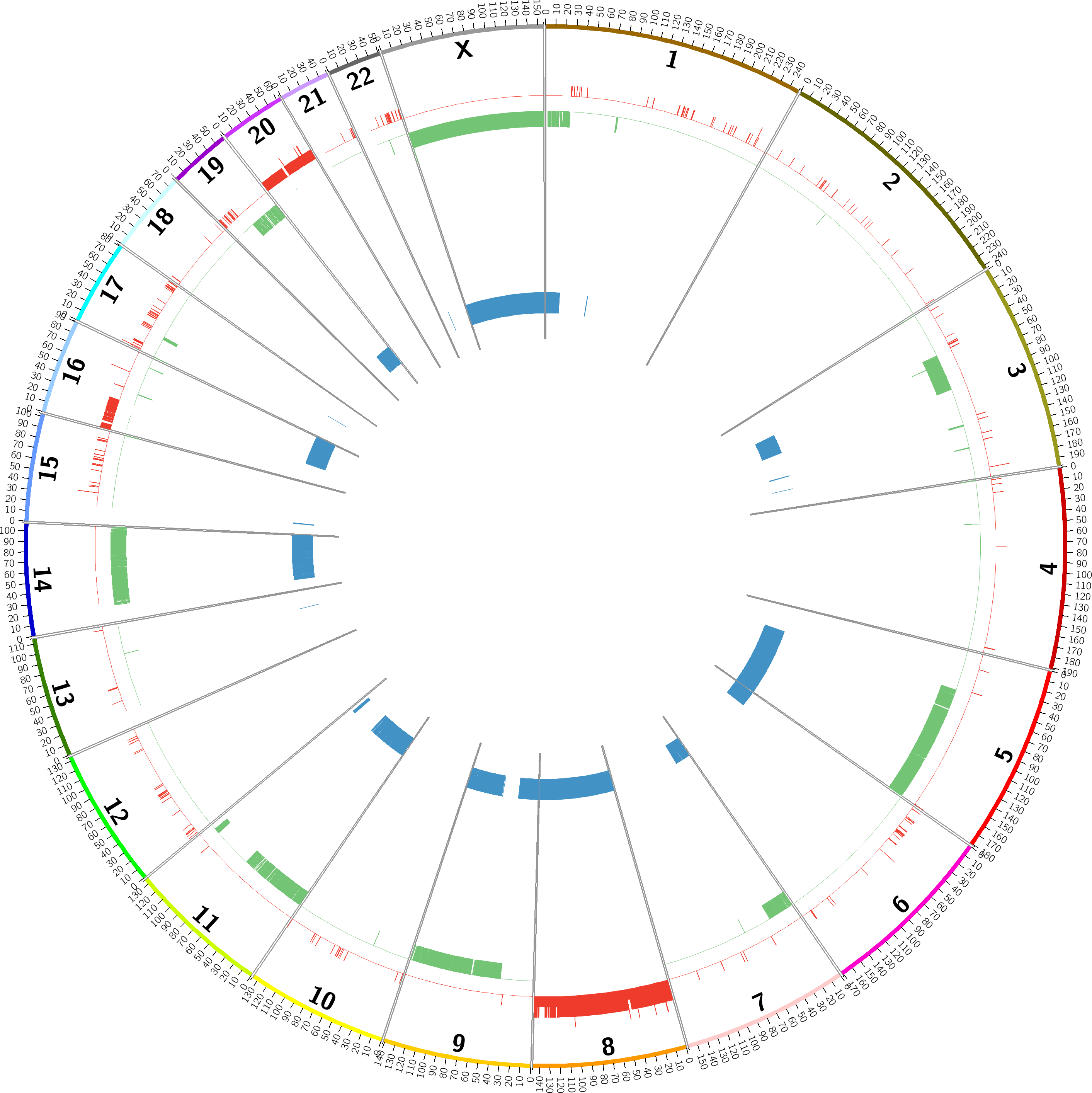
COPY NUMBER VARIANTS: GENOME DETAILS
CNVS OF POTENTIAL CLINICAL RELEVANCE
No copy number variants of clinical relevance were found from the analysis.
CNVS OF PROGNOSTIC OR DIAGNOSTIC RELEVANCE
No copy number variants of prognostic or diagnostic relevance were found from the analysis.
CNVS OF BIOLOGICAL RELEVANCE
| Gene | Copy Change | LOH State | CNV State | Chr:band | CNV Start | CNV End | Size (Mb) | Expression (RPKM) | Fold Change vs. average |
SARC %ile |
|---|---|---|---|---|---|---|---|---|---|---|
| APC | -1 | NLOH | Loss | 5:q22.2 | 111520402 | 112184695 | 1 | 2.46 | -2.25 | 36 |
COMMONLY AMPLIFIED ONCOGENES WITH COPY GAINS
| Gene | Copy Change | LOH State | CNV State | Chr:band | CNV Start | CNV End | Size (Mb) | Expression (RPKM) | Fold Change vs. average | SARC %ile |
|---|---|---|---|---|---|---|---|---|---|---|
| AURKA | +1 | HET | Gain | 20:q13.2-q13.33 | 52545359 | 62821928 | 10.28 | 13.28 | 4.55 | 73 |
| CCND2 | +1 | HET | Gain | 12:p13.32 | 4373073 | 4441890 | 0.07 | 100.32 | 3.86 | 98 |
| CDK4 | +1 | HET | Gain | 12:p13.3-q14.1 | 58075229 | 58392623 | 0.32 | 67.73 | 3.13 | 14 |
| FGFR1 | +2 | ALOH | Gain | 8:p11.23-p11.22 | 37597019 | 39231982 | 1.63 | 6.99 | -1.43 | 1 |
HOMOZYGOUSLY DELETED TUMOUR SUPPRESSORS
No homozygous deleted tumour suppressors were found from the analysis.
HIGHLY EXPRESSED ONCOGENES WITH COPY GAINS
| Gene | Copy Change | LOH State | CNV State | Chr:band | CNV Start | CNV End | Size (Mb) | Expression (RPKM) | Fold Change vs. average | SARC %ile |
|---|---|---|---|---|---|---|---|---|---|---|
| CCND | +1 | HET | Gain | 12:q13.32 | 4373073 | 4441890 | 0 | 100.32 | 3.86 | 98 |
| HOXD11 | +1 | HET | Gain | 2:q31.1 | 176936995 | 177010532 | 0 | 16.52 | 13.52 | 94 |
| HOXD13 | +1 | HET | Gain | 2:q31.1 | 176936995 | 177010532 | 0 | 59.93 | 49.76 | 100 |
| HSP90AB1 | +1 | HET | Gain | 6:p21.1 | 44205590 | 44315036 | 0 | 1494.2 | 3.25 | 99 |
| MYC | +2 | ALOH | Gain | 8:p24.21 | 128648140 | 128846228 | 0 | 60.13 | 2.67 | 86 |
| TOP1 | +1 | HET | Gain | 20:q11.21-q13.13 | 30169985 | 48766874 | 19 | 108.32 | 3.17 | 100 |
LOWLY EXPRESSED TUMOUR SUPPRESSORS WITH COPY LOSSES
| Gene | Copy Change | LOH State | CNV State | Chr:band | CNV Start | CNV End | Size (Mb) | Expression (RPKM) | Fold Change vs. average | SARC %ile |
|---|---|---|---|---|---|---|---|---|---|---|
| GPC3 | -1 | DLOH | Loss | X:p22.33-q28 | 182546 | 155260410 | 155.08 | 0.26 | -19.69 | 14 |
| KDM5C | -1 | DLOH | Loss | X:p22.33-q28 | 182546 | 155260410 | 155.08 | 2.83 | -2.56 | 0 |
| WT1 | -1 | DLOH | Loss | 11:p13 | 31270391 | 33703585 | 2.43 | 0.01 | -4.93 | 14 |
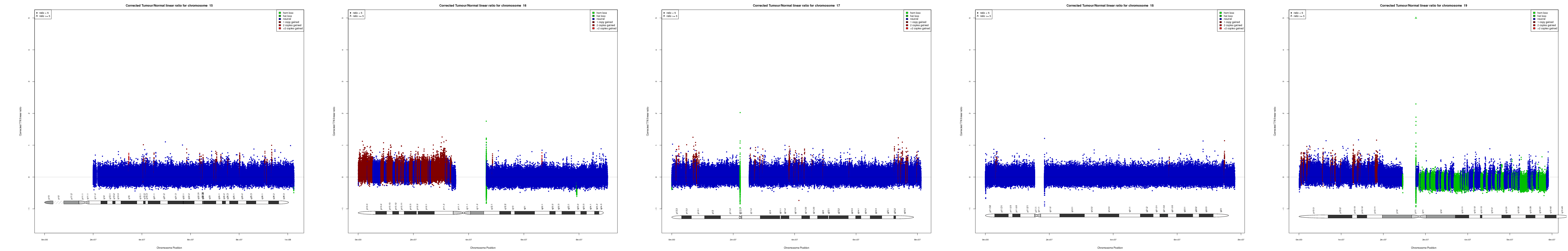
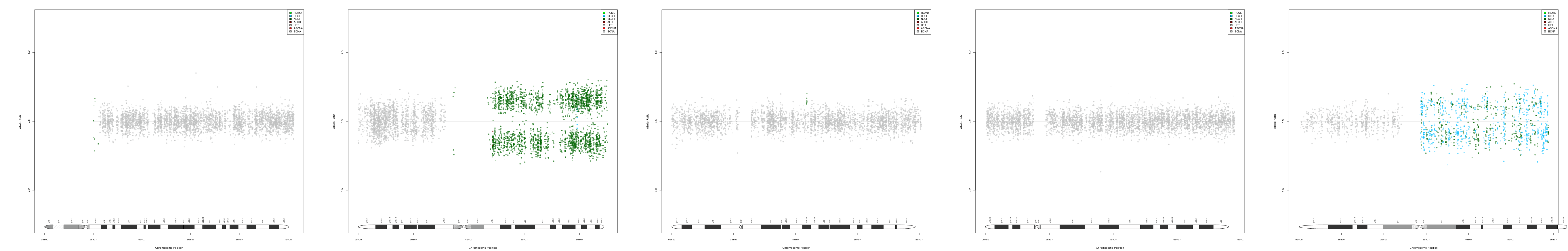
CNV AND LOH FIGURES
| Chr A | Chr 1 | Chr 2 | Chr 3 | Chr 4 |
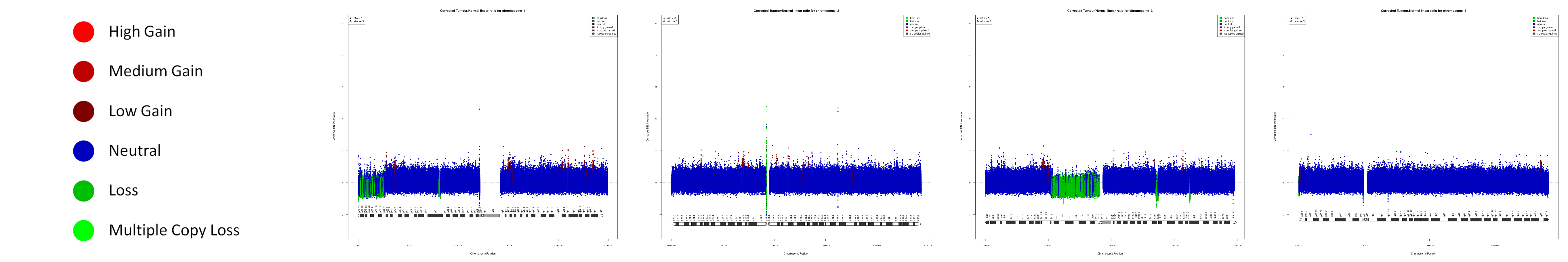 |
||||
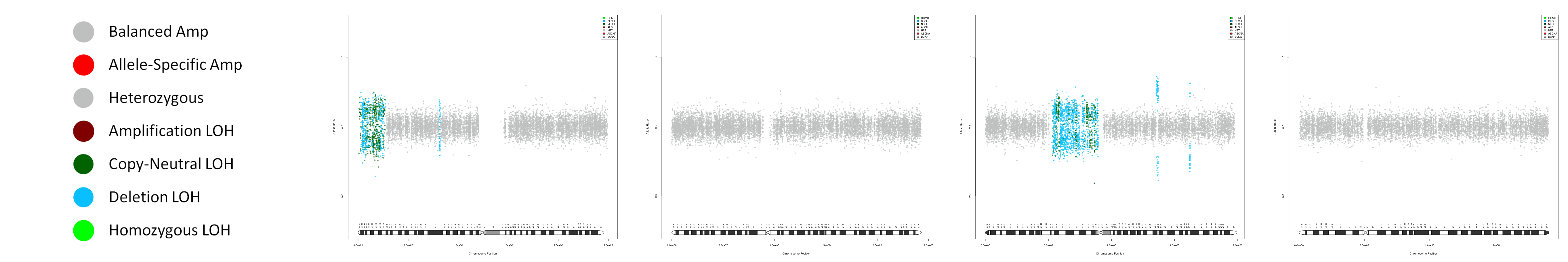 |
||||
| Chr 5 | Chr 6 | Chr 7 | Chr 8 | Chr 9 |
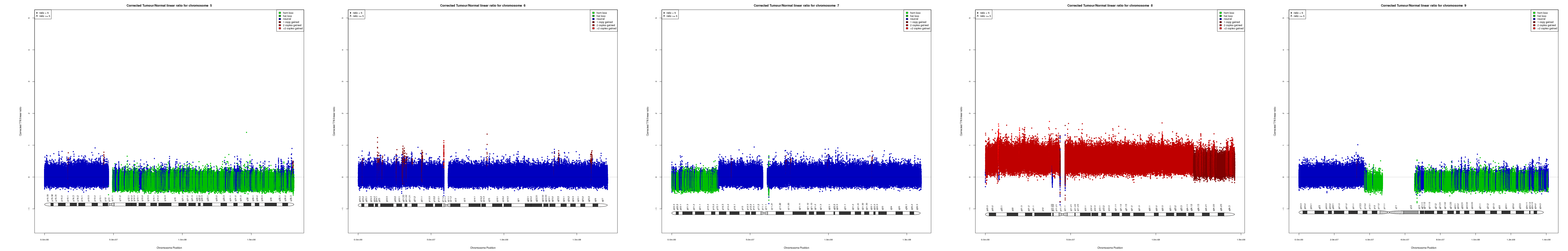 |
||||
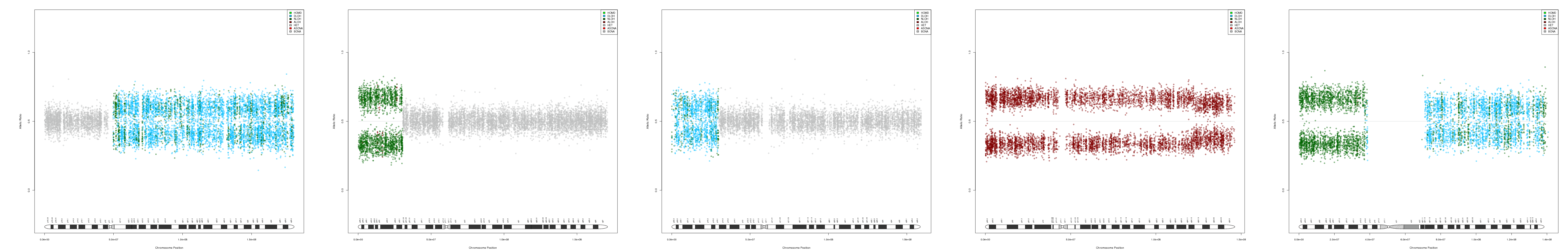 |
||||
| Chr 10 | Chr 11 | Chr 12 | Chr 13 | Chr 14 |
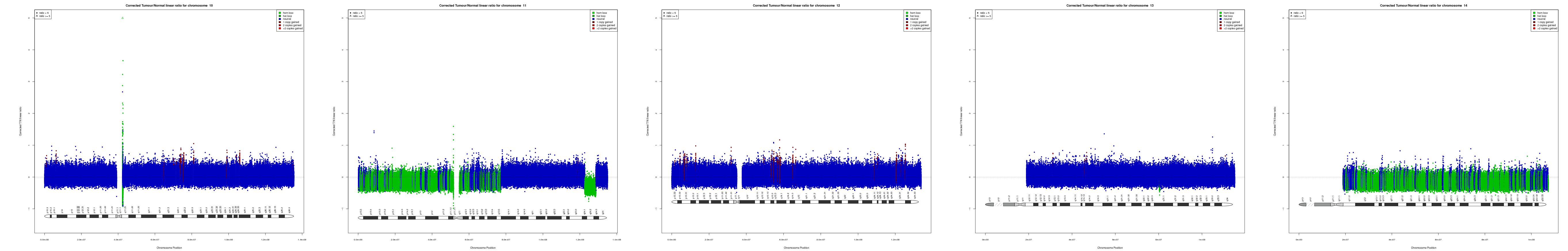 |
||||
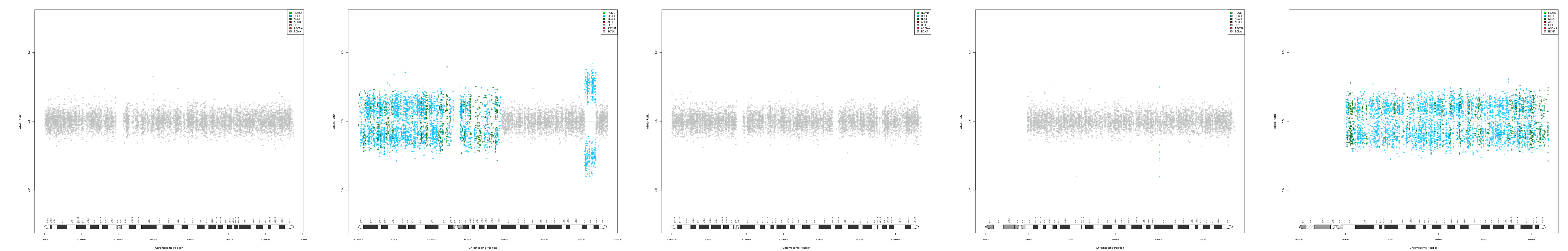 |
||||
| Chr 15 | Chr 16 | Chr 17 | Chr 18 | Chr 19 |
 |
||||
 |
||||