The following is a project page for Siu, C. (2017) Characterization of the normal thyroid reference epigenome (Master's thesis) under the supervision of Dr. Steven JM Jones.
The human thyroid, necessary for normal growth and development, is a relatively homogeneous tissue with a simple function, to produce and secrete appropriate levels of thyroid hormone for the regulation of metabolism in every cell of the human body. The accurate assessment of abnormal thyroids for different individuals is challenging and a fundamental understanding of the normal thyroid is therefore needed.
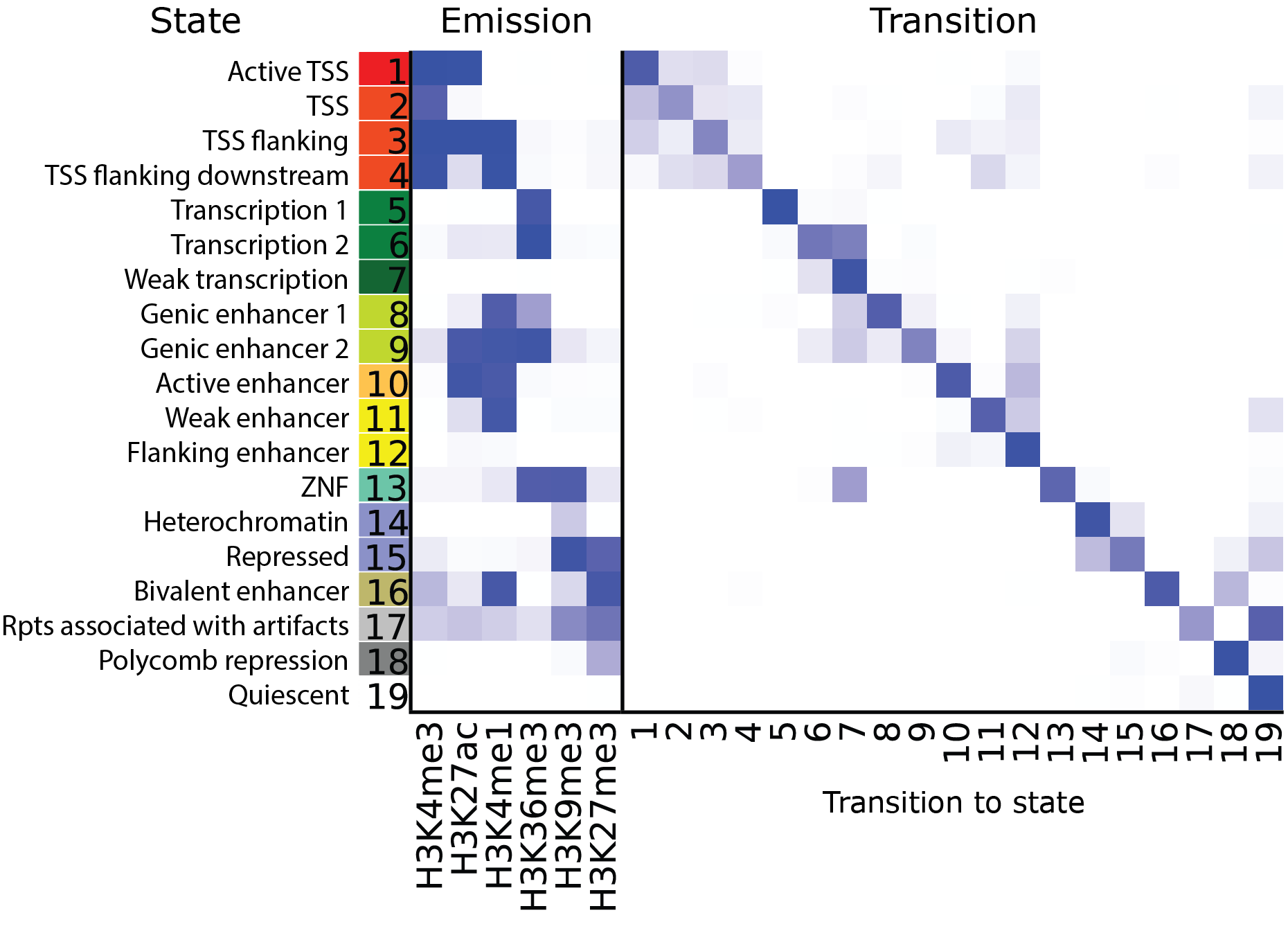
One way to characterize the normal thyroid is to study its epigenome and matched transcriptome. In this study, we compare the consistency of chromatin state annotations across the epigenomes from the grossly uninvolved tumour-adjacent thyroid tissue of four human individuals using ChIP-seq and RNA-seq. We examine 4 activating (H3K4me1, H3K4me3, H3K27ac, H3K36me3) and 2 repressing (H3K9me3, H3K27me3) histone modifications, identify chromatin states using a hidden Markov model (ChromHMM), produce a new metric for model selection, and establish epigenomic maps of 19 chromatin states.

Note: The input data and methods for training, model selection, and state labelling are described in [1].
| Track | Date last updated | UCSC | Data |
|---|---|---|---|
| ChromHMM state consensus and 19-state overview track | 2017-01-11 | Browser | Download |
| FindER consensus | 2017-01-11 | Browser | Download* |
| ChIP-seq overlay of histone modifications per sample | 2017-01-11 | Browser | ** |
| ChIP-seq overlap of samples per histone modification | 2017-01-11 | Browser | ** |
|
* Peak calls using MACS2 is also available ** Available/sourced from CEEHRC |
|||
The data presented in this work is a publicly funded data resource. You are free to use the data supplied by or through this web site in your scientific analysis. Please see our Open Access Data Release Policy page for more information. If you publish or use this data, you are required to attribute the original source of the data. Please cite [1] and acknowledge CEEHRC as a data source.
Aligned RNA-seq and ChIP-seq bam files are also available at http://www.epigenomes.ca/data-release/ and were provided by the Canadian Epigenetics, Environment, and Health Research Consortium (CEEHRC) initiative funded by the Canadian Institutes of Health Research (CIHR), Genome BC, and Genome Quebec.